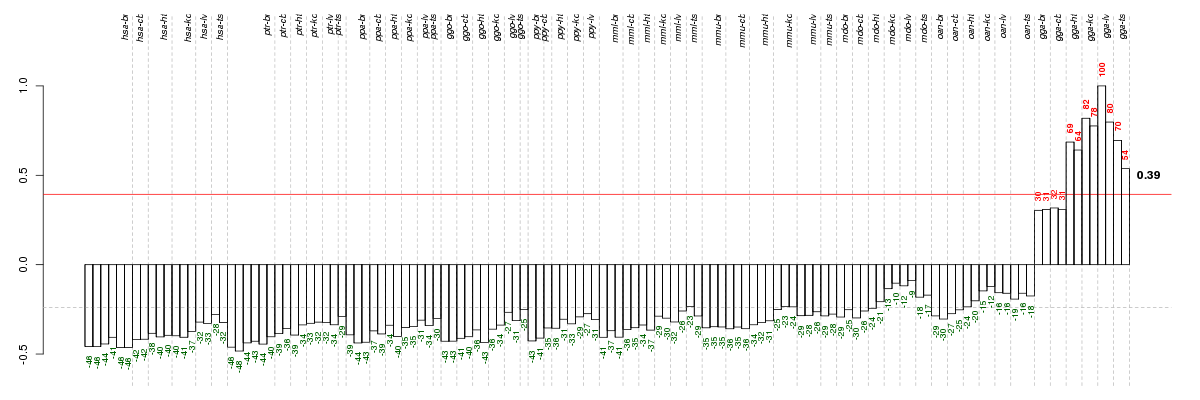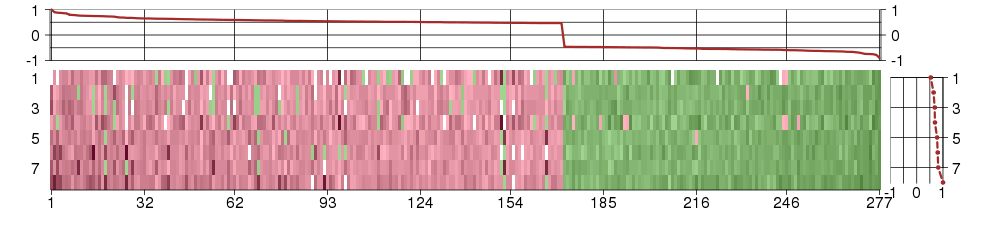

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
DNA binding
Any molecular function by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
NA
ABCA12ATP-binding cassette, sub-family A (ABC1), member 12 (ENSG00000144452), score: 0.73 ABLIM3actin binding LIM protein family, member 3 (ENSG00000173210), score: -0.46 ACAD9acyl-CoA dehydrogenase family, member 9 (ENSG00000177646), score: 0.7 ADAT1adenosine deaminase, tRNA-specific 1 (ENSG00000065457), score: 0.54 ADAT2adenosine deaminase, tRNA-specific 2, TAD2 homolog (S. cerevisiae) (ENSG00000189007), score: 0.63 AGFG1ArfGAP with FG repeats 1 (ENSG00000173744), score: 0.51 AMHanti-Mullerian hormone (ENSG00000104899), score: 0.54 ANO10anoctamin 10 (ENSG00000160746), score: -0.56 AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.55 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: 0.57 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (ENSG00000113108), score: -0.9 ARHGEF7Rho guanine nucleotide exchange factor (GEF) 7 (ENSG00000102606), score: -0.57 ARID3BAT rich interactive domain 3B (BRIGHT-like) (ENSG00000179361), score: 0.48 ARL3ADP-ribosylation factor-like 3 (ENSG00000138175), score: -0.62 ARNTaryl hydrocarbon receptor nuclear translocator (ENSG00000143437), score: 0.65 ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: -0.58 ATP7AATPase, Cu++ transporting, alpha polypeptide (ENSG00000165240), score: 0.62 BAHCC1BAH domain and coiled-coil containing 1 (ENSG00000171282), score: 0.49 BPGM2,3-bisphosphoglycerate mutase (ENSG00000172331), score: -0.6 BRD4bromodomain containing 4 (ENSG00000141867), score: 0.62 BRWD3bromodomain and WD repeat domain containing 3 (ENSG00000165288), score: 0.62 C12orf65chromosome 12 open reading frame 65 (ENSG00000130921), score: 0.55 C13orf39chromosome 13 open reading frame 39 (ENSG00000139780), score: 0.86 C14orf179chromosome 14 open reading frame 179 (ENSG00000119650), score: -0.49 C16orf42chromosome 16 open reading frame 42 (ENSG00000007520), score: -0.49 C18orf22chromosome 18 open reading frame 22 (ENSG00000101546), score: -0.48 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.64 C1orf55chromosome 1 open reading frame 55 (ENSG00000143751), score: 0.64 C20orf79chromosome 20 open reading frame 79 (ENSG00000132631), score: 0.65 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.6 C2orf65chromosome 2 open reading frame 65 (ENSG00000159374), score: 0.5 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.74 CAPN2calpain 2, (m/II) large subunit (ENSG00000162909), score: -0.57 CCDC47coiled-coil domain containing 47 (ENSG00000108588), score: -0.53 CD28CD28 molecule (ENSG00000178562), score: 0.62 CD40LGCD40 ligand (ENSG00000102245), score: 0.72 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.46 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 1 CDR2Lcerebellar degeneration-related protein 2-like (ENSG00000109089), score: -0.52 CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: -0.64 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.73 CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.68 CINPcyclin-dependent kinase 2 interacting protein (ENSG00000100865), score: -0.5 CMAScytidine monophosphate N-acetylneuraminic acid synthetase (ENSG00000111726), score: -0.64 CPEcarboxypeptidase E (ENSG00000109472), score: -0.57 CRISP1cysteine-rich secretory protein 1 (ENSG00000124812), score: 0.48 CRY1cryptochrome 1 (photolyase-like) (ENSG00000008405), score: 0.53 CRYBA1crystallin, beta A1 (ENSG00000108255), score: 0.53 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.54 CXXC5CXXC finger 5 (ENSG00000171604), score: -0.49 CYBASC3cytochrome b, ascorbate dependent 3 (ENSG00000162144), score: 0.47 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.52 DDX24DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 (ENSG00000089737), score: -0.55 DENND5ADENN/MADD domain containing 5A (ENSG00000184014), score: -0.54 DFFBDNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) (ENSG00000169598), score: 0.63 DHX29DEAH (Asp-Glu-Ala-His) box polypeptide 29 (ENSG00000067248), score: -0.6 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: -0.74 DIAPH2diaphanous homolog 2 (Drosophila) (ENSG00000147202), score: 0.53 DMAP1DNA methyltransferase 1 associated protein 1 (ENSG00000178028), score: -0.49 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: 0.51 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (ENSG00000168259), score: -0.47 DNMT3ADNA (cytosine-5-)-methyltransferase 3 alpha (ENSG00000119772), score: 0.52 EEDembryonic ectoderm development (ENSG00000074266), score: -0.53 EIF4EBP1eukaryotic translation initiation factor 4E binding protein 1 (ENSG00000187840), score: 0.47 EPS15L1epidermal growth factor receptor pathway substrate 15-like 1 (ENSG00000127527), score: -0.48 EREGepiregulin (ENSG00000124882), score: 0.67 ERGIC3ERGIC and golgi 3 (ENSG00000125991), score: -0.58 ERI1exoribonuclease 1 (ENSG00000104626), score: 0.57 ERLIN2ER lipid raft associated 2 (ENSG00000147475), score: 0.48 ESR2estrogen receptor 2 (ER beta) (ENSG00000140009), score: 0.58 ETV3ets variant 3 (ENSG00000117036), score: 0.65 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.51 FAM105Bfamily with sequence similarity 105, member B (ENSG00000154124), score: -0.47 FAM109Bfamily with sequence similarity 109, member B (ENSG00000177096), score: 0.8 FAM166Afamily with sequence similarity 166, member A (ENSG00000188163), score: -0.47 FANCAFanconi anemia, complementation group A (ENSG00000187741), score: -0.51 FBXL18F-box and leucine-rich repeat protein 18 (ENSG00000155034), score: 0.78 FBXL20F-box and leucine-rich repeat protein 20 (ENSG00000108306), score: 0.59 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.55 FIG4FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) (ENSG00000112367), score: -0.56 FNBP4formin binding protein 4 (ENSG00000109920), score: 0.61 FNDC7fibronectin type III domain containing 7 (ENSG00000143107), score: 0.59 FOXP2forkhead box P2 (ENSG00000128573), score: 0.55 FTSJD2FtsJ methyltransferase domain containing 2 (ENSG00000137200), score: -0.48 GALNT5UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) (ENSG00000136542), score: 0.54 GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: -0.57 GEMIN4gem (nuclear organelle) associated protein 4 (ENSG00000179409), score: 0.48 GNAI3guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 (ENSG00000065135), score: 0.52 GOPCgolgi-associated PDZ and coiled-coil motif containing (ENSG00000047932), score: -0.47 GORABgolgin, RAB6-interacting (ENSG00000120370), score: 0.52 GPR1G protein-coupled receptor 1 (ENSG00000183671), score: 0.5 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.47 GPR149G protein-coupled receptor 149 (ENSG00000174948), score: 0.53 GPR64G protein-coupled receptor 64 (ENSG00000173698), score: 0.48 GXYLT2glucoside xylosyltransferase 2 (ENSG00000172986), score: 0.49 HDAC8histone deacetylase 8 (ENSG00000147099), score: 0.48 HDHD3haloacid dehalogenase-like hydrolase domain containing 3 (ENSG00000119431), score: 0.51 HELZhelicase with zinc finger (ENSG00000198265), score: 0.47 HMG20Ahigh-mobility group 20A (ENSG00000140382), score: -0.57 HMGXB3HMG box domain containing 3 (ENSG00000113716), score: -0.49 HNRNPA0heterogeneous nuclear ribonucleoprotein A0 (ENSG00000177733), score: -0.48 HPGDShematopoietic prostaglandin D synthase (ENSG00000163106), score: 0.87 HSF4heat shock transcription factor 4 (ENSG00000102878), score: -0.47 ICOSinducible T-cell co-stimulator (ENSG00000163600), score: 0.76 IKZF3IKAROS family zinc finger 3 (Aiolos) (ENSG00000161405), score: 0.56 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.74 IL15interleukin 15 (ENSG00000164136), score: 0.51 IL22RA2interleukin 22 receptor, alpha 2 (ENSG00000164485), score: 0.67 IL7Rinterleukin 7 receptor (ENSG00000168685), score: 0.51 IMPG2interphotoreceptor matrix proteoglycan 2 (ENSG00000081148), score: 0.78 INO80CINO80 complex subunit C (ENSG00000153391), score: -0.49 INPP5Kinositol polyphosphate-5-phosphatase K (ENSG00000132376), score: -0.6 INPPL1inositol polyphosphate phosphatase-like 1 (ENSG00000165458), score: -0.55 INTS2integrator complex subunit 2 (ENSG00000108506), score: 0.49 IRF8interferon regulatory factor 8 (ENSG00000140968), score: 0.64 KBTBD8kelch repeat and BTB (POZ) domain containing 8 (ENSG00000163376), score: 0.47 KIAA0368KIAA0368 (ENSG00000136813), score: -0.57 KIAA1737KIAA1737 (ENSG00000198894), score: -0.53 KIRRELkin of IRRE like (Drosophila) (ENSG00000183853), score: 0.5 KLC4kinesin light chain 4 (ENSG00000137171), score: 0.51 KLF8Kruppel-like factor 8 (ENSG00000102349), score: 0.57 KLHDC1kelch domain containing 1 (ENSG00000197776), score: 0.59 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.5 KLHL28kelch-like 28 (Drosophila) (ENSG00000179454), score: 0.6 KREMEN1kringle containing transmembrane protein 1 (ENSG00000183762), score: -0.62 KRT9keratin 9 (ENSG00000171403), score: 0.5 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.69 LASS6LAG1 homolog, ceramide synthase 6 (ENSG00000172292), score: -0.59 LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (ENSG00000131023), score: 0.6 LCMT1leucine carboxyl methyltransferase 1 (ENSG00000205629), score: -0.71 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.63 LRP11low density lipoprotein receptor-related protein 11 (ENSG00000120256), score: -0.48 LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.66 LSM4LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) (ENSG00000130520), score: -0.64 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: -0.53 MAPKSP1MAPK scaffold protein 1 (ENSG00000109270), score: 0.5 MBD3methyl-CpG binding domain protein 3 (ENSG00000071655), score: -0.56 MBNL3muscleblind-like 3 (Drosophila) (ENSG00000076770), score: 0.52 MPLmyeloproliferative leukemia virus oncogene (ENSG00000117400), score: 0.59 MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.58 NAT10N-acetyltransferase 10 (GCN5-related) (ENSG00000135372), score: -0.48 NCSTNnicastrin (ENSG00000162736), score: -0.47 NLRC3NLR family, CARD domain containing 3 (ENSG00000167984), score: 0.52 NOL6nucleolar protein family 6 (RNA-associated) (ENSG00000165271), score: -0.49 NR0B1nuclear receptor subfamily 0, group B, member 1 (ENSG00000169297), score: 0.55 NRF1nuclear respiratory factor 1 (ENSG00000106459), score: 0.52 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: -0.51 NUSAP1nucleolar and spindle associated protein 1 (ENSG00000137804), score: 0.5 OLFML2Aolfactomedin-like 2A (ENSG00000185585), score: 0.67 ORC2Lorigin recognition complex, subunit 2-like (yeast) (ENSG00000115942), score: -0.57 ORC5Lorigin recognition complex, subunit 5-like (yeast) (ENSG00000164815), score: -0.63 OTUD1OTU domain containing 1 (ENSG00000165312), score: 0.52 PAIP1poly(A) binding protein interacting protein 1 (ENSG00000172239), score: -0.65 PANK3pantothenate kinase 3 (ENSG00000120137), score: 0.51 PANX3pannexin 3 (ENSG00000154143), score: 0.85 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.75 PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (ENSG00000140694), score: -0.66 PATZ1POZ (BTB) and AT hook containing zinc finger 1 (ENSG00000100105), score: -0.6 PCGF2polycomb group ring finger 2 (ENSG00000056661), score: -0.53 PDCD1LG2programmed cell death 1 ligand 2 (ENSG00000197646), score: 0.55 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.5 PDIK1LPDLIM1 interacting kinase 1 like (ENSG00000175087), score: 0.48 PDX1pancreatic and duodenal homeobox 1 (ENSG00000139515), score: 0.6 PHLDA3pleckstrin homology-like domain, family A, member 3 (ENSG00000174307), score: -0.47 PICALMphosphatidylinositol binding clathrin assembly protein (ENSG00000073921), score: -0.51 PIK3CGphosphoinositide-3-kinase, catalytic, gamma polypeptide (ENSG00000105851), score: 0.65 PLCG1phospholipase C, gamma 1 (ENSG00000124181), score: -0.55 PLRG1pleiotropic regulator 1 (PRL1 homolog, Arabidopsis) (ENSG00000171566), score: -0.75 POF1Bpremature ovarian failure, 1B (ENSG00000124429), score: 0.59 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (ENSG00000100479), score: 0.73 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (ENSG00000125630), score: 0.58 POMGNT1protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase (ENSG00000085998), score: -0.48 PPME1protein phosphatase methylesterase 1 (ENSG00000214517), score: -0.47 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta (ENSG00000175470), score: -0.49 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta (ENSG00000112640), score: -0.52 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.54 PRKAG3protein kinase, AMP-activated, gamma 3 non-catalytic subunit (ENSG00000115592), score: 0.74 PRPF40APRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) (ENSG00000196504), score: 0.48 PTDSS1phosphatidylserine synthase 1 (ENSG00000156471), score: -0.47 QDPRquinoid dihydropteridine reductase (ENSG00000151552), score: -0.51 QPCTLglutaminyl-peptide cyclotransferase-like (ENSG00000011478), score: 0.61 QTRTD1queuine tRNA-ribosyltransferase domain containing 1 (ENSG00000151576), score: 0.57 RAB19RAB19, member RAS oncogene family (ENSG00000146955), score: 0.58 RAB22ARAB22A, member RAS oncogene family (ENSG00000124209), score: -0.51 RAB27ARAB27A, member RAS oncogene family (ENSG00000069974), score: 0.53 RAB3GAP1RAB3 GTPase activating protein subunit 1 (catalytic) (ENSG00000115839), score: -0.63 RAB4ARAB4A, member RAS oncogene family (ENSG00000168118), score: -0.62 RANBP10RAN binding protein 10 (ENSG00000141084), score: 0.51 RAPGEF6Rap guanine nucleotide exchange factor (GEF) 6 (ENSG00000158987), score: 0.47 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.49 RBP1retinol binding protein 1, cellular (ENSG00000114115), score: -0.6 RELv-rel reticuloendotheliosis viral oncogene homolog (avian) (ENSG00000162924), score: 0.66 RESTRE1-silencing transcription factor (ENSG00000084093), score: 0.49 RHAGRh-associated glycoprotein (ENSG00000112077), score: 0.64 RHOBTB3Rho-related BTB domain containing 3 (ENSG00000164292), score: -0.57 RPIAribose 5-phosphate isomerase A (ENSG00000153574), score: 0.75 RXFP3relaxin/insulin-like family peptide receptor 3 (ENSG00000182631), score: 0.47 SAV1salvador homolog 1 (Drosophila) (ENSG00000151748), score: 0.48 SDF4stromal cell derived factor 4 (ENSG00000078808), score: -0.61 SEPT2septin 2 (ENSG00000168385), score: -0.78 SESN1sestrin 1 (ENSG00000080546), score: 0.47 SFPQsplicing factor proline/glutamine-rich (ENSG00000116560), score: -0.55 SHISA2shisa homolog 2 (Xenopus laevis) (ENSG00000180730), score: 0.5 SLBPstem-loop binding protein (ENSG00000163950), score: -0.59 SLC18A1solute carrier family 18 (vesicular monoamine), member 1 (ENSG00000036565), score: 0.55 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: 0.47 SLC25A38solute carrier family 25, member 38 (ENSG00000144659), score: -0.52 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.87 SMOsmoothened homolog (Drosophila) (ENSG00000128602), score: 0.47 SNX29sorting nexin 29 (ENSG00000048471), score: -0.48 SNX33sorting nexin 33 (ENSG00000173548), score: 0.53 SOS1son of sevenless homolog 1 (Drosophila) (ENSG00000115904), score: 0.5 SOS2son of sevenless homolog 2 (Drosophila) (ENSG00000100485), score: 0.47 SPHK1sphingosine kinase 1 (ENSG00000176170), score: 0.49 SPINK4serine peptidase inhibitor, Kazal type 4 (ENSG00000122711), score: 0.89 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (ENSG00000184402), score: -0.56 SSNA1Sjogren syndrome nuclear autoantigen 1 (ENSG00000176101), score: -0.56 SSR1signal sequence receptor, alpha (ENSG00000124783), score: 0.49 STK24serine/threonine kinase 24 (ENSG00000102572), score: -0.55 SYDE2synapse defective 1, Rho GTPase, homolog 2 (C. elegans) (ENSG00000097096), score: 0.59 TAAR1trace amine associated receptor 1 (ENSG00000146399), score: 0.57 TADA2Btranscriptional adaptor 2B (ENSG00000173011), score: -0.54 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (ENSG00000172660), score: -0.55 TAF3TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa (ENSG00000165632), score: 0.76 TAF5TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa (ENSG00000148835), score: 0.57 TALDO1transaldolase 1 (ENSG00000177156), score: -0.48 TAS1R1taste receptor, type 1, member 1 (ENSG00000173662), score: 0.47 TATDN1TatD DNase domain containing 1 (ENSG00000147687), score: 0.59 THOC2THO complex 2 (ENSG00000125676), score: 0.53 TIRAPtoll-interleukin 1 receptor (TIR) domain containing adaptor protein (ENSG00000150455), score: 0.52 TLX1T-cell leukemia homeobox 1 (ENSG00000107807), score: 0.48 TM7SF4transmembrane 7 superfamily member 4 (ENSG00000164935), score: 0.52 TMCO7transmembrane and coiled-coil domains 7 (ENSG00000103047), score: 0.54 TMEM199transmembrane protein 199 (ENSG00000244045), score: 0.74 TMEM26transmembrane protein 26 (ENSG00000196932), score: 0.48 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.61 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.64 TMEM87Atransmembrane protein 87A (ENSG00000103978), score: 0.55 TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.59 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: -0.66 TNFRSF11Atumor necrosis factor receptor superfamily, member 11a, NFKB activator (ENSG00000141655), score: 0.75 TOMM40Ltranslocase of outer mitochondrial membrane 40 homolog (yeast)-like (ENSG00000158882), score: 0.47 TP63tumor protein p63 (ENSG00000073282), score: 0.61 TRAF3IP3TRAF3 interacting protein 3 (ENSG00000009790), score: 0.49 TRAT1T cell receptor associated transmembrane adaptor 1 (ENSG00000163519), score: 0.55 TRPA1transient receptor potential cation channel, subfamily A, member 1 (ENSG00000104321), score: 0.58 TSHBthyroid stimulating hormone, beta (ENSG00000134200), score: 0.54 TTC1tetratricopeptide repeat domain 1 (ENSG00000113312), score: -0.48 TTLL1tubulin tyrosine ligase-like family, member 1 (ENSG00000100271), score: -0.64 TUBB1tubulin, beta 1 (ENSG00000101162), score: 0.65 UBA2ubiquitin-like modifier activating enzyme 2 (ENSG00000126261), score: -0.55 UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.62 UBE2Kubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) (ENSG00000078140), score: -0.48 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: -0.62 UBL7ubiquitin-like 7 (bone marrow stromal cell-derived) (ENSG00000138629), score: -0.58 UFSP2UFM1-specific peptidase 2 (ENSG00000109775), score: -0.55 UNC119unc-119 homolog (C. elegans) (ENSG00000109103), score: -0.68 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (ENSG00000175073), score: 0.5 VPS33Bvacuolar protein sorting 33 homolog B (yeast) (ENSG00000184056), score: 0.57 VTI1Avesicle transport through interaction with t-SNAREs homolog 1A (yeast) (ENSG00000151532), score: 0.5 WDFY4WDFY family member 4 (ENSG00000128815), score: 0.51 WDR41WD repeat domain 41 (ENSG00000164253), score: -0.75 WDR73WD repeat domain 73 (ENSG00000177082), score: 0.52 XPR1xenotropic and polytropic retrovirus receptor 1 (ENSG00000143324), score: 0.63 YPEL5yippee-like 5 (Drosophila) (ENSG00000119801), score: -0.47 ZBTB2zinc finger and BTB domain containing 2 (ENSG00000181472), score: 0.49 ZBTB33zinc finger and BTB domain containing 33 (ENSG00000177485), score: 0.54 ZBTB37zinc finger and BTB domain containing 37 (ENSG00000185278), score: 0.51 ZBTB8Bzinc finger and BTB domain containing 8B (ENSG00000215897), score: 0.75 ZC3H12Azinc finger CCCH-type containing 12A (ENSG00000163874), score: 0.47 ZFHX3zinc finger homeobox 3 (ENSG00000140836), score: 0.61 ZMYM2zinc finger, MYM-type 2 (ENSG00000121741), score: -0.58 ZNF704zinc finger protein 704 (ENSG00000164684), score: 0.68 ZNF828zinc finger protein 828 (ENSG00000198824), score: 0.53 ZP1zona pellucida glycoprotein 1 (sperm receptor) (ENSG00000149506), score: 0.54
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_ts_m2_ca1 | gga | ts | m | 2 |
| gga_ht_f_ca1 | gga | ht | f | _ |
| gga_ht_m_ca1 | gga | ht | m | _ |
| gga_ts_m1_ca1 | gga | ts | m | 1 |
| gga_kd_f_ca1 | gga | kd | f | _ |
| gga_lv_f_ca1 | gga | lv | f | _ |
| gga_kd_m_ca1 | gga | kd | m | _ |
| gga_lv_m_ca1 | gga | lv | m | _ |