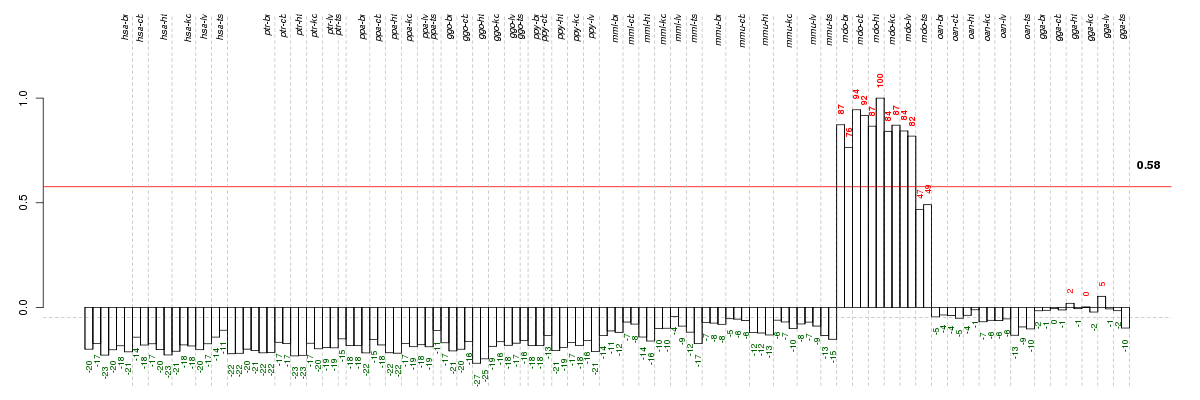

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
protein amino acid lipidation
The covalent or non-covalent attachment of lipid moieties to an amino acid in a protein.
GPI anchor metabolic process
The chemical reactions and pathways involving glycosylphosphatidylinositol anchors, molecular mechanisms for attaching membrane proteins to the lipid bilayer of cell membranes. Structurally they consist of a molecule of phosphatidylinositol to which is linked, via the C-6 hydroxyl of the inositol, a carbohydrate chain. This chain is in turn linked to the protein through an ethanolamine phosphate moiety, the amino group of which is in amide linkage with the C-terminal carboxyl of the protein chain, the phosphate group being esterified to the C-6 hydroxyl of the terminal mannose of the core carbohydrate chain.
GPI anchor biosynthetic process
The chemical reactions and pathways resulting in the formation of a glycosylphosphatidylinositol (GPI) anchor that attaches some membrane proteins to the lipid bilayer of the cell membrane. The phosphatidylinositol moiety is linked via the C-6 hydroxyl residue of inositol to a carbohydrate chain which is itself linked to the protein via an ethanolamine phosphate moiety, its amino group forming an amide linkage with the C-terminal carboxyl of the protein. Some GPI anchors have variants on this canonical linkage.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
phospholipid metabolic process
The chemical reactions and pathways involving phospholipids, any lipid containing phosphoric acid as a mono- or diester.
glycerophospholipid metabolic process
The chemical reactions and pathways involving glycerophospholipids, any derivative of glycerophosphate that contains at least one O-acyl, O-alkyl, or O-alkenyl group attached to the glycerol residue.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
lipid biosynthetic process
The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
phospholipid biosynthetic process
The chemical reactions and pathways resulting in the formation of phospholipids, any lipid containing phosphoric acid as a mono- or diester.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
attachment of GPI anchor to protein
A transamidation reaction that results in the cleavage of the polypeptide chain and the concomitant transfer of the GPI anchor to the newly formed carboxy-terminal amino acid of the anchored protein. The cleaved C-terminal contains the C-terminal GPI signal sequence of the newly synthesized polypeptide chain.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
organophosphate metabolic process
The chemical reactions and pathways involving organophosphates, any phosphate-containing organic compound.
phosphoinositide metabolic process
The chemical reactions and pathways involving phosphoinositides, any of a class of glycerophospholipids in which the phosphatidyl group is esterified to the hydroxyl group of inositol. They are important constituents of cell membranes.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
lipoprotein metabolic process
The chemical reactions and pathways involving any conjugated, water-soluble protein in which the nonprotein moiety consists of a lipid or lipids.
lipoprotein biosynthetic process
The chemical reactions and pathways resulting in the formation of any conjugated, water-soluble protein in which the nonprotein moiety consists of a lipid or lipids.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
macromolecule modification
The covalent alteration of one or more monomeric units in a polypeptide, polynucleotide, polysaccharide, or other biological macromolecule, resulting in a change in its properties.
post-translational protein modification
The covalent alteration of one or more amino acids occurring in a protein after the protein has been completely translated and released from the ribosome.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
glycerolipid biosynthetic process
The chemical reactions and pathways resulting in the formation of glycerolipids, any lipid with a glycerol backbone.
glycerophospholipid biosynthetic process
The chemical reactions and pathways resulting in the formation of glycerophospholipids, any derivative of glycerophosphate that contains at least one O-acyl, O-alkyl, or O-alkenyl group attached to the glycerol residue.
glycerolipid metabolic process
The chemical reactions and pathways involving glycerolipids, any lipid with a glycerol backbone. Diacylglycerol and phosphatidate are key lipid intermediates of glycerolipid biosynthesis.
phosphoinositide biosynthetic process
The chemical reactions and pathways resulting in the formation of phosphoinositides, any of a class of glycerophospholipids in which the phosphatidyl group is esterified to the hydroxyl group of inositol.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
phospholipid biosynthetic process
The chemical reactions and pathways resulting in the formation of phospholipids, any lipid containing phosphoric acid as a mono- or diester.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
glycerolipid biosynthetic process
The chemical reactions and pathways resulting in the formation of glycerolipids, any lipid with a glycerol backbone.
phospholipid biosynthetic process
The chemical reactions and pathways resulting in the formation of phospholipids, any lipid containing phosphoric acid as a mono- or diester.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
phospholipid metabolic process
The chemical reactions and pathways involving phospholipids, any lipid containing phosphoric acid as a mono- or diester.
lipid biosynthetic process
The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
glycerophospholipid biosynthetic process
The chemical reactions and pathways resulting in the formation of glycerophospholipids, any derivative of glycerophosphate that contains at least one O-acyl, O-alkyl, or O-alkenyl group attached to the glycerol residue.
glycerophospholipid biosynthetic process
The chemical reactions and pathways resulting in the formation of glycerophospholipids, any derivative of glycerophosphate that contains at least one O-acyl, O-alkyl, or O-alkenyl group attached to the glycerol residue.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
lipoprotein biosynthetic process
The chemical reactions and pathways resulting in the formation of any conjugated, water-soluble protein in which the nonprotein moiety consists of a lipid or lipids.
glycerophospholipid metabolic process
The chemical reactions and pathways involving glycerophospholipids, any derivative of glycerophosphate that contains at least one O-acyl, O-alkyl, or O-alkenyl group attached to the glycerol residue.
glycerolipid biosynthetic process
The chemical reactions and pathways resulting in the formation of glycerolipids, any lipid with a glycerol backbone.
protein amino acid lipidation
The covalent or non-covalent attachment of lipid moieties to an amino acid in a protein.
phosphoinositide biosynthetic process
The chemical reactions and pathways resulting in the formation of phosphoinositides, any of a class of glycerophospholipids in which the phosphatidyl group is esterified to the hydroxyl group of inositol.
GPI anchor biosynthetic process
The chemical reactions and pathways resulting in the formation of a glycosylphosphatidylinositol (GPI) anchor that attaches some membrane proteins to the lipid bilayer of the cell membrane. The phosphatidylinositol moiety is linked via the C-6 hydroxyl residue of inositol to a carbohydrate chain which is itself linked to the protein via an ethanolamine phosphate moiety, its amino group forming an amide linkage with the C-terminal carboxyl of the protein. Some GPI anchors have variants on this canonical linkage.
GPI anchor biosynthetic process
The chemical reactions and pathways resulting in the formation of a glycosylphosphatidylinositol (GPI) anchor that attaches some membrane proteins to the lipid bilayer of the cell membrane. The phosphatidylinositol moiety is linked via the C-6 hydroxyl residue of inositol to a carbohydrate chain which is itself linked to the protein via an ethanolamine phosphate moiety, its amino group forming an amide linkage with the C-terminal carboxyl of the protein. Some GPI anchors have variants on this canonical linkage.
attachment of GPI anchor to protein
A transamidation reaction that results in the cleavage of the polypeptide chain and the concomitant transfer of the GPI anchor to the newly formed carboxy-terminal amino acid of the anchored protein. The cleaved C-terminal contains the C-terminal GPI signal sequence of the newly synthesized polypeptide chain.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
integral to endoplasmic reticulum membrane
Penetrating at least one phospholipid bilayer of an endoplasmic reticulum membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
organelle membrane
The lipid bilayer surrounding an organelle.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to endoplasmic reticulum membrane
Located in the endoplasmic reticulum membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to organelle membrane
Penetrating at least one phospholipid bilayer of an organelle membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
GPI-anchor transamidase complex
An enzyme complex which in humans and yeast consists of at least five proteins; for example, the complex contains GAA1, GPI8, PIG-S, PIG-U, and PIG-T in human, and Gaa1p, Gab1p, Gpi8p, Gpi16p, and Gpi17p in yeast. Catalyzes the posttranslational attachment of the carboxyl-terminus of a precursor protein to a GPI-anchor.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
GPI-anchor transamidase complex
An enzyme complex which in humans and yeast consists of at least five proteins; for example, the complex contains GAA1, GPI8, PIG-S, PIG-U, and PIG-T in human, and Gaa1p, Gab1p, Gpi8p, Gpi16p, and Gpi17p in yeast. Catalyzes the posttranslational attachment of the carboxyl-terminus of a precursor protein to a GPI-anchor.
intrinsic to endoplasmic reticulum membrane
Located in the endoplasmic reticulum membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
intrinsic to endoplasmic reticulum membrane
Located in the endoplasmic reticulum membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
GPI-anchor transamidase complex
An enzyme complex which in humans and yeast consists of at least five proteins; for example, the complex contains GAA1, GPI8, PIG-S, PIG-U, and PIG-T in human, and Gaa1p, Gab1p, Gpi8p, Gpi16p, and Gpi17p in yeast. Catalyzes the posttranslational attachment of the carboxyl-terminus of a precursor protein to a GPI-anchor.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
integral to endoplasmic reticulum membrane
Penetrating at least one phospholipid bilayer of an endoplasmic reticulum membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
integral to organelle membrane
Penetrating at least one phospholipid bilayer of an organelle membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
GPI-anchor transamidase complex
An enzyme complex which in humans and yeast consists of at least five proteins; for example, the complex contains GAA1, GPI8, PIG-S, PIG-U, and PIG-T in human, and Gaa1p, Gab1p, Gpi8p, Gpi16p, and Gpi17p in yeast. Catalyzes the posttranslational attachment of the carboxyl-terminus of a precursor protein to a GPI-anchor.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
GPI-anchor transamidase activity
Catalysis of the formation of the linkage between a protein and a glycosylphosphatidylinositol anchor. The reaction probably occurs by subjecting a peptide bond to nucleophilic attack by the amino group of ethanolamine-GPI, transferring the protein from a signal peptide to the GPI anchor.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00563 | 6.738e-03 | 0.5507 | 5 | 11 | Glycosylphosphatidylinositol(GPI)-anchor biosynthesis |
AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: -0.62 AATFapoptosis antagonizing transcription factor (ENSG00000108270), score: -0.61 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (ENSG00000160179), score: -0.49 ACANaggrecan (ENSG00000157766), score: 0.53 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: 0.54 ACBD6acyl-CoA binding domain containing 6 (ENSG00000135847), score: -0.49 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.61 ACTL6Aactin-like 6A (ENSG00000136518), score: 0.45 ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.9 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.74 AHSA1AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) (ENSG00000100591), score: 0.45 AHSA2AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) (ENSG00000173209), score: -0.48 AICDAactivation-induced cytidine deaminase (ENSG00000111732), score: 0.44 ALKBH1alkB, alkylation repair homolog 1 (E. coli) (ENSG00000100601), score: 0.54 ALKBH2alkB, alkylation repair homolog 2 (E. coli) (ENSG00000189046), score: 0.71 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.73 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.62 ANKMY2ankyrin repeat and MYND domain containing 2 (ENSG00000106524), score: 0.55 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.49 ANKRD42ankyrin repeat domain 42 (ENSG00000137494), score: -0.54 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: 0.45 ARHGDIGRho GDP dissociation inhibitor (GDI) gamma (ENSG00000167930), score: -0.55 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 0.5 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.65 ASB1ankyrin repeat and SOCS box-containing 1 (ENSG00000065802), score: 0.69 ATG16L1ATG16 autophagy related 16-like 1 (S. cerevisiae) (ENSG00000085978), score: 0.68 ATG4AATG4 autophagy related 4 homolog A (S. cerevisiae) (ENSG00000101844), score: 0.46 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: 0.65 ATMINATM interactor (ENSG00000166454), score: -0.47 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.67 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.45 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.84 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: -0.51 BCL2L13BCL2-like 13 (apoptosis facilitator) (ENSG00000099968), score: -0.55 BCL7AB-cell CLL/lymphoma 7A (ENSG00000110987), score: 0.49 BLVRAbiliverdin reductase A (ENSG00000106605), score: -0.64 BTKBruton agammaglobulinemia tyrosine kinase (ENSG00000010671), score: 0.52 C10orf137chromosome 10 open reading frame 137 (ENSG00000107938), score: 0.45 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.6 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.64 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.65 C13orf31chromosome 13 open reading frame 31 (ENSG00000179630), score: 0.8 C14orf101chromosome 14 open reading frame 101 (ENSG00000070269), score: -0.57 C14orf45chromosome 14 open reading frame 45 (ENSG00000119636), score: -0.48 C15orf29chromosome 15 open reading frame 29 (ENSG00000134152), score: -0.55 C15orf60chromosome 15 open reading frame 60 (ENSG00000183324), score: 0.5 C17orf75chromosome 17 open reading frame 75 (ENSG00000108666), score: 0.55 C1orf103chromosome 1 open reading frame 103 (ENSG00000121931), score: -0.52 C1orf123chromosome 1 open reading frame 123 (ENSG00000162384), score: 0.7 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.74 C22orf13chromosome 22 open reading frame 13 (ENSG00000138867), score: 0.45 C2CD2C2 calcium-dependent domain containing 2 (ENSG00000157617), score: -0.52 C3orf23chromosome 3 open reading frame 23 (ENSG00000179152), score: 0.54 C3orf39chromosome 3 open reading frame 39 (ENSG00000144647), score: 0.68 C3orf59chromosome 3 open reading frame 59 (ENSG00000180611), score: 0.57 C6orf186chromosome 6 open reading frame 186 (ENSG00000053328), score: 0.45 C6orf57chromosome 6 open reading frame 57 (ENSG00000154079), score: 0.57 C6orf64chromosome 6 open reading frame 64 (ENSG00000112167), score: -0.51 C6orf89chromosome 6 open reading frame 89 (ENSG00000198663), score: 0.48 C9orf102chromosome 9 open reading frame 102 (ENSG00000182150), score: -0.59 C9orf103chromosome 9 open reading frame 103 (ENSG00000148057), score: -0.47 CA13carbonic anhydrase XIII (ENSG00000185015), score: 0.46 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.58 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (ENSG00000078699), score: 0.48 CCDC6coiled-coil domain containing 6 (ENSG00000108091), score: 0.5 CCDC77coiled-coil domain containing 77 (ENSG00000120647), score: -0.58 CCDC82coiled-coil domain containing 82 (ENSG00000149231), score: -0.64 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.79 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: -0.57 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.82 CDK10cyclin-dependent kinase 10 (ENSG00000185324), score: -0.53 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (ENSG00000115816), score: -0.5 CENPTcentromere protein T (ENSG00000102901), score: -0.5 CHD1Lchromodomain helicase DNA binding protein 1-like (ENSG00000131778), score: -0.62 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: -0.65 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.51 CLTBclathrin, light chain B (ENSG00000175416), score: -0.55 CNTFciliary neurotrophic factor (ENSG00000186660), score: 0.46 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.88 CPSF6cleavage and polyadenylation specific factor 6, 68kDa (ENSG00000111605), score: 0.47 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.56 CSTF1cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa (ENSG00000101138), score: -0.51 CTDP1CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 (ENSG00000060069), score: -0.5 CWF19L2CWF19-like 2, cell cycle control (S. pombe) (ENSG00000152404), score: -0.53 CYLDcylindromatosis (turban tumor syndrome) (ENSG00000083799), score: -0.64 CYTBcytochrome b (ENSG00000198727), score: -1 CYTH1cytohesin 1 (ENSG00000108669), score: -0.66 DCTDdCMP deaminase (ENSG00000129187), score: -0.52 DCTN5dynactin 5 (p25) (ENSG00000166847), score: -0.52 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (ENSG00000109184), score: -0.55 DDX1DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (ENSG00000079785), score: 0.63 DDX47DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (ENSG00000213782), score: -0.61 DDX51DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 (ENSG00000185163), score: -0.52 DENND5BDENN/MADD domain containing 5B (ENSG00000170456), score: 0.49 DERL3Der1-like domain family, member 3 (ENSG00000099958), score: 0.54 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: -0.57 DHRS7Bdehydrogenase/reductase (SDR family) member 7B (ENSG00000109016), score: -0.47 DPH5DPH5 homolog (S. cerevisiae) (ENSG00000117543), score: 0.48 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (ENSG00000105865), score: -0.47 EEA1early endosome antigen 1 (ENSG00000102189), score: 0.47 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (ENSG00000172071), score: 0.5 ELMO3engulfment and cell motility 3 (ENSG00000102890), score: 0.49 EPB41L4Aerythrocyte membrane protein band 4.1 like 4A (ENSG00000129595), score: -0.5 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.74 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.68 EXD2exonuclease 3'-5' domain containing 2 (ENSG00000081177), score: -0.63 EXOC3exocyst complex component 3 (ENSG00000180104), score: -0.51 EXOSC9exosome component 9 (ENSG00000123737), score: 0.57 FAM118Bfamily with sequence similarity 118, member B (ENSG00000197798), score: -0.52 FAM160A2family with sequence similarity 160, member A2 (ENSG00000051009), score: 0.45 FAM20Bfamily with sequence similarity 20, member B (ENSG00000116199), score: 0.46 FAM26Efamily with sequence similarity 26, member E (ENSG00000178033), score: 0.44 FAM40Bfamily with sequence similarity 40, member B (ENSG00000128578), score: 0.52 FAM73Afamily with sequence similarity 73, member A (ENSG00000180488), score: 0.64 FAM82A2family with sequence similarity 82, member A2 (ENSG00000137824), score: 0.65 FBXO15F-box protein 15 (ENSG00000141665), score: 0.52 FBXO22F-box protein 22 (ENSG00000167196), score: 0.65 FBXO3F-box protein 3 (ENSG00000110429), score: 0.6 FRA10AC1fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 (ENSG00000148690), score: -0.63 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.81 GARSglycyl-tRNA synthetase (ENSG00000106105), score: 0.62 GBGT1globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 (ENSG00000148288), score: 0.57 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.62 GLIPR2GLI pathogenesis-related 2 (ENSG00000122694), score: -0.57 GLT8D1glycosyltransferase 8 domain containing 1 (ENSG00000016864), score: -0.57 GMNNgeminin, DNA replication inhibitor (ENSG00000112312), score: -0.52 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: -0.49 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.69 GRAMD2GRAM domain containing 2 (ENSG00000175318), score: 0.46 GTF3Ageneral transcription factor IIIA (ENSG00000122034), score: -0.62 HAGHhydroxyacylglutathione hydrolase (ENSG00000063854), score: -0.52 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (ENSG00000153187), score: 0.52 HPRT1hypoxanthine phosphoribosyltransferase 1 (ENSG00000165704), score: 0.5 HSF1heat shock transcription factor 1 (ENSG00000185122), score: -0.48 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.75 IKBIPIKBKB interacting protein (ENSG00000166130), score: 0.62 INVSinversin (ENSG00000119509), score: -0.6 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.96 KBTBD8kelch repeat and BTB (POZ) domain containing 8 (ENSG00000163376), score: 0.52 KCTD5potassium channel tetramerisation domain containing 5 (ENSG00000167977), score: -0.66 KCTD6potassium channel tetramerisation domain containing 6 (ENSG00000168301), score: 0.48 KIAA0090KIAA0090 (ENSG00000127463), score: 0.5 KIAA0146KIAA0146 (ENSG00000164808), score: -0.54 KIAA0174KIAA0174 (ENSG00000182149), score: -0.6 KIAA1432KIAA1432 (ENSG00000107036), score: 0.5 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.53 L3MBTL2l(3)mbt-like 2 (Drosophila) (ENSG00000100395), score: 0.48 LAMP2lysosomal-associated membrane protein 2 (ENSG00000005893), score: -0.55 LEPRE1leucine proline-enriched proteoglycan (leprecan) 1 (ENSG00000117385), score: 0.53 LOC100134387similar to DEAH (Asp-Glu-Ala-His) box polypeptide 40 (ENSG00000108406), score: -0.57 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: 0.57 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: -0.69 MCM8minichromosome maintenance complex component 8 (ENSG00000125885), score: -0.5 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (ENSG00000082212), score: -0.69 MED1mediator complex subunit 1 (ENSG00000125686), score: 0.54 MEMO1mediator of cell motility 1 (ENSG00000162961), score: -0.6 MESDC1mesoderm development candidate 1 (ENSG00000140406), score: 0.56 METRNLmeteorin, glial cell differentiation regulator-like (ENSG00000176845), score: -0.62 MGAT4Amannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A (ENSG00000071073), score: -0.51 MID2midline 2 (ENSG00000080561), score: 0.52 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.67 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: -0.53 MTERFD1MTERF domain containing 1 (ENSG00000156469), score: 0.47 MTHFD2Lmethylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (ENSG00000163738), score: 0.45 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: -0.54 MTMR14myotubularin related protein 14 (ENSG00000163719), score: 0.47 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.68 NADSYN1NAD synthetase 1 (ENSG00000172890), score: 0.47 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.59 NARFnuclear prelamin A recognition factor (ENSG00000141562), score: -0.64 NARG2NMDA receptor regulated 2 (ENSG00000128915), score: -0.54 ND2MTND2 (ENSG00000198763), score: -0.96 ND6NADH dehydrogenase, subunit 6 (complex I) (ENSG00000198695), score: -0.58 NEK8NIMA (never in mitosis gene a)- related kinase 8 (ENSG00000160602), score: 0.46 NFIAnuclear factor I/A (ENSG00000162599), score: 0.53 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: 0.71 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.96 NRBF2nuclear receptor binding factor 2 (ENSG00000148572), score: 0.53 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.74 NUP85nucleoporin 85kDa (ENSG00000125450), score: 0.52 OCIAD1OCIA domain containing 1 (ENSG00000109180), score: -0.55 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: -0.49 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.5 OTOL1otolin 1 homolog (zebrafish) (ENSG00000182447), score: 0.69 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: -0.51 PARP1poly (ADP-ribose) polymerase 1 (ENSG00000143799), score: -0.48 PARS2prolyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000162396), score: 0.46 PCID2PCI domain containing 2 (ENSG00000126226), score: -0.58 PDE12phosphodiesterase 12 (ENSG00000174840), score: 0.51 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.65 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.61 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (ENSG00000170525), score: -0.61 PGRprogesterone receptor (ENSG00000082175), score: 0.65 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.64 PHF21APHD finger protein 21A (ENSG00000135365), score: 0.65 PHF5APHD finger protein 5A (ENSG00000100410), score: 0.54 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.73 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.75 PIGPphosphatidylinositol glycan anchor biosynthesis, class P (ENSG00000185808), score: 0.44 PIGSphosphatidylinositol glycan anchor biosynthesis, class S (ENSG00000087111), score: 0.76 PIGUphosphatidylinositol glycan anchor biosynthesis, class U (ENSG00000101464), score: 0.46 PLDNpallidin homolog (mouse) (ENSG00000104164), score: -0.5 PNPLA6patatin-like phospholipase domain containing 6 (ENSG00000032444), score: 0.5 PODNpodocan (ENSG00000174348), score: 0.64 POLR3Apolymerase (RNA) III (DNA directed) polypeptide A, 155kDa (ENSG00000148606), score: -0.48 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (ENSG00000161980), score: 0.47 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta (ENSG00000175470), score: 0.46 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.55 PRKACBprotein kinase, cAMP-dependent, catalytic, beta (ENSG00000142875), score: -0.53 PSEN1presenilin 1 (ENSG00000080815), score: -0.48 PTRH2peptidyl-tRNA hydrolase 2 (ENSG00000141378), score: -0.6 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.64 QSER1glutamine and serine rich 1 (ENSG00000060749), score: 0.49 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.47 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.53 RALAv-ral simian leukemia viral oncogene homolog A (ras related) (ENSG00000006451), score: 0.51 RBM6RNA binding motif protein 6 (ENSG00000004534), score: -0.53 RBPJLrecombination signal binding protein for immunoglobulin kappa J region-like (ENSG00000124232), score: 0.64 RCAN1regulator of calcineurin 1 (ENSG00000159200), score: -0.53 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.49 RFXANKregulatory factor X-associated ankyrin-containing protein (ENSG00000064490), score: 0.46 RG9MTD1RNA (guanine-9-) methyltransferase domain containing 1 (ENSG00000174173), score: -0.6 RGS2regulator of G-protein signaling 2, 24kDa (ENSG00000116741), score: -0.49 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.84 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.69 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.51 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.55 RTN4IP1reticulon 4 interacting protein 1 (ENSG00000130347), score: 0.46 SAP30LSAP30-like (ENSG00000164576), score: 0.7 SEC22ASEC22 vesicle trafficking protein homolog A (S. cerevisiae) (ENSG00000121542), score: -0.49 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.58 SEH1LSEH1-like (S. cerevisiae) (ENSG00000085415), score: 0.45 SELSselenoprotein S (ENSG00000131871), score: -0.51 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.57 SFRS8splicing factor, arginine/serine-rich 8 (suppressor-of-white-apricot homolog, Drosophila) (ENSG00000061936), score: 0.71 SIN3BSIN3 homolog B, transcription regulator (yeast) (ENSG00000127511), score: -0.52 SIRT1sirtuin 1 (ENSG00000096717), score: 0.46 SLC15A4solute carrier family 15, member 4 (ENSG00000139370), score: 0.56 SLC25A3solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 (ENSG00000075415), score: 0.47 SLC25A38solute carrier family 25, member 38 (ENSG00000144659), score: -0.47 SLC30A9solute carrier family 30 (zinc transporter), member 9 (ENSG00000014824), score: -0.51 SLC35B3solute carrier family 35, member B3 (ENSG00000124786), score: -0.65 SLC38A1solute carrier family 38, member 1 (ENSG00000111371), score: -0.53 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.96 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (ENSG00000100335), score: 0.52 SNX13sorting nexin 13 (ENSG00000071189), score: 0.63 SNX8sorting nexin 8 (ENSG00000106266), score: -0.56 SPG20spastic paraplegia 20 (Troyer syndrome) (ENSG00000133104), score: -0.64 SRPX2sushi-repeat-containing protein, X-linked 2 (ENSG00000102359), score: 0.55 STAMBPSTAM binding protein (ENSG00000124356), score: 0.66 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (ENSG00000040341), score: -0.56 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.64 STX16syntaxin 16 (ENSG00000124222), score: -0.49 STX8syntaxin 8 (ENSG00000170310), score: -0.48 SYT12synaptotagmin XII (ENSG00000173227), score: 0.47 TCF3transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) (ENSG00000071564), score: 0.55 TCP11L2t-complex 11 (mouse)-like 2 (ENSG00000166046), score: 0.5 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.74 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (ENSG00000151923), score: 0.58 TMCO6transmembrane and coiled-coil domains 6 (ENSG00000113119), score: 0.65 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.51 TMEM138transmembrane protein 138 (ENSG00000149483), score: 0.47 TMEM18transmembrane protein 18 (ENSG00000151353), score: -0.7 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.89 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.45 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.67 TPH2tryptophan hydroxylase 2 (ENSG00000139287), score: 0.48 TRAF1TNF receptor-associated factor 1 (ENSG00000056558), score: -0.52 TRAF3TNF receptor-associated factor 3 (ENSG00000131323), score: -0.5 TSC22D2TSC22 domain family, member 2 (ENSG00000196428), score: 0.56 TSNtranslin (ENSG00000211460), score: -0.59 TSPAN13tetraspanin 13 (ENSG00000106537), score: 0.49 TSPAN9tetraspanin 9 (ENSG00000011105), score: 0.45 TTC12tetratricopeptide repeat domain 12 (ENSG00000149292), score: -0.52 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.59 TUSC3tumor suppressor candidate 3 (ENSG00000104723), score: 0.49 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.96 UBN2ubinuclein 2 (ENSG00000157741), score: -0.5 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.61 UCK1uridine-cytidine kinase 1 (ENSG00000130717), score: 0.51 UNC50unc-50 homolog (C. elegans) (ENSG00000115446), score: -0.69 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: 0.64 UTP11LUTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) (ENSG00000183520), score: 0.49 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.6 VAC14Vac14 homolog (S. cerevisiae) (ENSG00000103043), score: 0.48 VPS33Bvacuolar protein sorting 33 homolog B (yeast) (ENSG00000184056), score: 0.53 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (ENSG00000139722), score: 0.48 VPS39vacuolar protein sorting 39 homolog (S. cerevisiae) (ENSG00000166887), score: -0.48 WDR43WD repeat domain 43 (ENSG00000163811), score: 0.45 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: 0.49 ZBTB11zinc finger and BTB domain containing 11 (ENSG00000066422), score: -0.49 ZBTB43zinc finger and BTB domain containing 43 (ENSG00000169155), score: -0.48 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -1 ZFYVE27zinc finger, FYVE domain containing 27 (ENSG00000155256), score: 0.52 ZNF276zinc finger protein 276 (ENSG00000158805), score: 0.46 ZNF346zinc finger protein 346 (ENSG00000113761), score: -0.61 ZNF385Czinc finger protein 385C (ENSG00000187595), score: 0.47 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.73 ZNF800zinc finger protein 800 (ENSG00000048405), score: 0.46 ZRANB3zinc finger, RAN-binding domain containing 3 (ENSG00000121988), score: -0.49 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.7
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_br_f_ca1 | mdo | br | f | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |