Previous module |
Next module
Module #512, TG: 2, TC: 2, 157 probes, 157 Entrez genes, 6 conditions


HELP
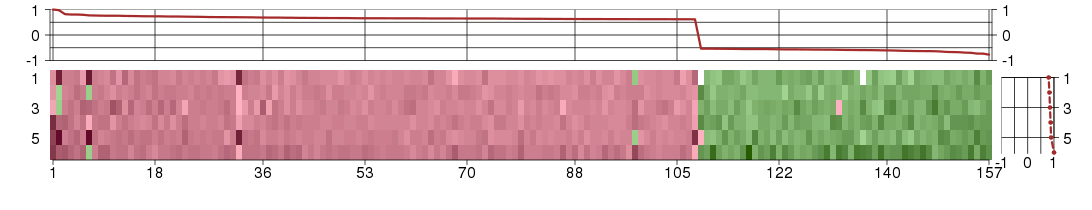
The image plot shows the color-coded level of gene expression, for the
genes and conditions in a given transcription module. The genes are on
the horizontal, the conditions on the vertical axis.
The genes are ordered according to their ISA gene scores, similarly
the conditions are ordered according to their condition scores. The
score of a gene means the «degree of inclusion» in
the module: a high score gene is essential in the module.
Condition scores can also be negative, that means that the genes of
the module are all down-regulated in the condition. Here the absolute
value of the score gives the «degree of inclusion».
The plots above and beside the expression matrix show the gene scores
and condition scores, respectively.
Note that the plot is interactive, you can see the name of the gene
and condition under the mouse cursor.
The expression matrix was normalized to have mean zero and standard
deviation one for every gene separately across all conditions
(i.e. not just for the conditions in the module).
— Click on the Help button again to close this help window.

Under-expression is coded with green,
over-expression with red color.
Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Biological processes
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for biological processes.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Cellular Components
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for cellular components.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Molecular Function
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for molecular function.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
GO BP test for over-representation
HELP
List of all enriched GO categories (biological processes), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0006401 | 2.469e-02 | 0.6769 | 5
ABCE1, MTPAP, PAN3, XRN1, XRN2 | 23 | RNA catabolic process |
Help |
Hide |
Top
Help |
Show |
Top
GO CC test for over-representation
HELP
List of all enriched GO categories (cellular components), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0008076 | 3.103e-02 | 1.338 | 6
KCNF1, KCNG1, KCNG3, KCNQ5, KCNS1, KCNS2 | 46 | voltage-gated potassium channel complex |
| GO:0034705 | 3.103e-02 | 1.338 | 6
KCNF1, KCNG1, KCNG3, KCNQ5, KCNS1, KCNS2 | 46 | potassium channel complex |
| GO:0044450 | 4.634e-02 | 0.6401 | 4
CETN3, EXOC7, NEDD1, ROCK1 | 22 | microtubule organizing center part |
Help |
Hide |
Top
Help |
Show |
Top
GO MF test for over-representation
HELP
List of all enriched GO categories (molecular function), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0004930 | 1.720e-02 | 3.254 | 11
BAI1, CHRM3, GPR123, GPR176, HRH3, HTR2A, LGR4, MC4R, NTSR1, OPN4, OPRL1 | 120 | G-protein coupled receptor activity |
| GO:0004428 | 2.411e-02 | 0.4067 | 4
ITPK1, ITPKA, PIK3C2A, PIK3R2 | 15 | inositol or phosphatidylinositol kinase activity |
| GO:0004683 | 4.793e-02 | 0.244 | 3
CAMK1G, CAMK2A, ITPKA | 9 | calmodulin-dependent protein kinase activity |
| GO:0008227 | 4.793e-02 | 0.244 | 3
CHRM3, HRH3, HTR2A | 9 | G-protein coupled amine receptor activity |
Help |
Hide |
Top
Help |
Show |
Top
KEGG Pathway test for over-representation
HELP
List of all enriched KEGG pathways, at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given KEGG pathway, just by chance.
- Count
is the number of genes in the module annotated with the given KEGG
pathway.
- Size is the total number of genes (in our universe)
annotated with the KEGG pathway.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
KEGG pathway.
Clicking on the KEGG identifiers takes you to the KEGG web site.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| 04070 | 3.750e-02 | 0.8106 | 5
DGKZ, ITPK1, ITPKA, PIK3C2A, PIK3R2 | 32 | Phosphatidylinositol signaling system |
Help |
Hide |
Top
Help |
Show |
Top
miRNA test for over-representation
HELP
List of all enriched miRNA families, at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module regulated by the given miRNA family, just by chance.
- Count
is the number of genes in the module regulated by the given miRNA
family.
- Size is the total number of genes (in our universe)
regulated with the given miRNA family.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
miRNA family.
The miRNA regulation data was taken from the TargetScan database.
(Only the conserved sites were used for the current analysis.)
Clicking on the miRNA names takes you to the TargetScan web site.
— Click on the Help button again to close this help window.
Help |
Hide |
Top
Help |
Show |
Top
Chromosome test for over-representation
HELP
List of all enriched Chromosomes, at the 0.05
p-value level.
The columns:
- ExpCount is the expected number of genes in the
module on the given chromosome, just by chance.
- Count
is the number of genes in the module on the given chromosome.
- Size is the total number of genes (in our universe)
on the given chromosome.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
chromosome.
— Click on the Help button again to close this help window.
HELP
A list of all genes in the current module, in alphabetical order. The
size of the text corresponds to the gene scores.
Text color indicates correlation and anti-correlation: genes of the
same color are correlated, both move up or down in the module samples
(listed below). Genes of different color are anti-correlated, they
have opposite behavior in the module samples.
Note, that text color of the individual genes should be interpreted
together with the coloring of the samples below. For
red samples,
red genes have a higher expression
(compared to the average gene expression level),
green genes have a lower
expression.
Green samples have opposite
behavior, in these red genes have a
lower expression, green genes
have a higher expression.
Note that some gene symbols may show up more than once, if many
probes match the same Entrez gene.
Genes with no Entrez mapping are given separately, with their
Affymetrics probe ID.
— Click on the Help button again to close this help window.
Entrez genes
ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: -0.62
ACTL6Aactin-like 6A (ENSG00000136518), score: -0.54
ADAMTS8ADAM metallopeptidase with thrombospondin type 1 motif, 8 (ENSG00000134917), score: 0.72
ANKRD24ankyrin repeat domain 24 (ENSG00000089847), score: 0.63
ARCactivity-regulated cytoskeleton-associated protein (ENSG00000198576), score: 0.68
ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: -0.59
ASB6ankyrin repeat and SOCS box-containing 6 (ENSG00000148331), score: 0.73
ATP13A2ATPase type 13A2 (ENSG00000159363), score: 0.65
BAI1brain-specific angiogenesis inhibitor 1 (ENSG00000181790), score: 0.64
BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (ENSG00000163930), score: 0.8
C13orf36chromosome 13 open reading frame 36 (ENSG00000180440), score: 0.66
C1orf55chromosome 1 open reading frame 55 (ENSG00000143751), score: -0.55
C8orf46chromosome 8 open reading frame 46 (ENSG00000169085), score: 0.65
C9orf16chromosome 9 open reading frame 16 (ENSG00000171159), score: 0.7
CA7carbonic anhydrase VII (ENSG00000168748), score: 0.65
CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 1
CAMK1Gcalcium/calmodulin-dependent protein kinase IG (ENSG00000008118), score: 0.67
CAMK2Acalcium/calmodulin-dependent protein kinase II alpha (ENSG00000070808), score: 0.65
CAPN9calpain 9 (ENSG00000135773), score: 0.72
CBLN4cerebellin 4 precursor (ENSG00000054803), score: 0.75
CD101CD101 molecule (ENSG00000134256), score: 0.7
CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.63
CELF5CUGBP, Elav-like family member 5 (ENSG00000161082), score: 0.62
CETN3centrin, EF-hand protein, 3 (ENSG00000153140), score: -0.59
CHRM3cholinergic receptor, muscarinic 3 (ENSG00000133019), score: 0.65
CHST1carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 (ENSG00000175264), score: 0.65
CNIH3cornichon homolog 3 (Drosophila) (ENSG00000143786), score: 0.65
COPB1coatomer protein complex, subunit beta 1 (ENSG00000129083), score: -0.6
DGCR2DiGeorge syndrome critical region gene 2 (ENSG00000070413), score: 0.68
DGKZdiacylglycerol kinase, zeta 104kDa (ENSG00000149091), score: 0.7
DLX1distal-less homeobox 1 (ENSG00000144355), score: 0.69
ELFN2extracellular leucine-rich repeat and fibronectin type III domain containing 2 (ENSG00000166897), score: 0.65
ELMOD2ELMO/CED-12 domain containing 2 (ENSG00000179387), score: -0.77
ENC1ectodermal-neural cortex 1 (with BTB-like domain) (ENSG00000171617), score: 0.71
EPHB6EPH receptor B6 (ENSG00000106123), score: 0.73
ETAA1Ewing tumor-associated antigen 1 (ENSG00000143971), score: -0.6
EXOC7exocyst complex component 7 (ENSG00000182473), score: 0.7
FAM19A1family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 (ENSG00000183662), score: 0.73
FAM5Bfamily with sequence similarity 5, member B (ENSG00000198797), score: 0.65
FBLN7fibulin 7 (ENSG00000144152), score: 0.74
FBXL16F-box and leucine-rich repeat protein 16 (ENSG00000127585), score: 0.62
FBXO8F-box protein 8 (ENSG00000164117), score: -0.59
FCHO2FCH domain only 2 (ENSG00000157107), score: -0.56
FERD3LFer3-like (Drosophila) (ENSG00000146618), score: 0.77
FEZF2FEZ family zinc finger 2 (ENSG00000153266), score: 0.75
FOXG1forkhead box G1 (ENSG00000176165), score: 0.71
GABPAGA binding protein transcription factor, alpha subunit 60kDa (ENSG00000154727), score: -0.64
GJD4gap junction protein, delta 4, 40.1kDa (ENSG00000177291), score: 0.62
GNB1guanine nucleotide binding protein (G protein), beta polypeptide 1 (ENSG00000078369), score: 0.65
GOLGA4golgin A4 (ENSG00000144674), score: -0.7
GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.82
GPR176G protein-coupled receptor 176 (ENSG00000166073), score: 0.66
GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: -0.57
HAT1histone acetyltransferase 1 (ENSG00000128708), score: -0.65
HRH3histamine receptor H3 (ENSG00000101180), score: 0.64
HTR2A5-hydroxytryptamine (serotonin) receptor 2A (ENSG00000102468), score: 0.76
IDEinsulin-degrading enzyme (ENSG00000119912), score: -0.57
INO80DINO80 complex subunit D (ENSG00000114933), score: -0.55
INTS8integrator complex subunit 8 (ENSG00000164941), score: -0.56
ITPK1inositol 1,3,4-triphosphate 5/6 kinase (ENSG00000100605), score: 0.64
ITPKAinositol 1,4,5-trisphosphate 3-kinase A (ENSG00000137825), score: 0.63
KCNF1potassium voltage-gated channel, subfamily F, member 1 (ENSG00000162975), score: 0.8
KCNG1potassium voltage-gated channel, subfamily G, member 1 (ENSG00000026559), score: 0.74
KCNG3potassium voltage-gated channel, subfamily G, member 3 (ENSG00000171126), score: 0.76
KCNQ5potassium voltage-gated channel, KQT-like subfamily, member 5 (ENSG00000185760), score: 0.67
KCNS1potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 (ENSG00000124134), score: 0.73
KCNS2potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 (ENSG00000156486), score: 0.63
KDM5Alysine (K)-specific demethylase 5A (ENSG00000073614), score: -0.57
KIAA0284KIAA0284 (ENSG00000099814), score: 0.66
KIAA1033KIAA1033 (ENSG00000136051), score: -0.57
KIRREL3kin of IRRE like 3 (Drosophila) (ENSG00000149571), score: 0.62
KLHDC3kelch domain containing 3 (ENSG00000124702), score: 0.63
LAPTM4Alysosomal protein transmembrane 4 alpha (ENSG00000068697), score: -0.54
LBRlamin B receptor (ENSG00000143815), score: -0.59
LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: -0.57
LIMK1LIM domain kinase 1 (ENSG00000106683), score: 0.62
LMO4LIM domain only 4 (ENSG00000143013), score: 0.7
LMTK2lemur tyrosine kinase 2 (ENSG00000164715), score: 0.64
LOC100132308solute carrier family 29 (nucleoside transporters), member 4 pseudogene (ENSG00000164638), score: 0.63
LPPR3lipid phosphate phosphatase-related protein type 3 (ENSG00000129951), score: 0.76
LPPR4lipid phosphate phosphatase-related protein type 4 (ENSG00000117600), score: 0.62
LRFN2leucine rich repeat and fibronectin type III domain containing 2 (ENSG00000156564), score: 0.76
LRTM2leucine-rich repeats and transmembrane domains 2 (ENSG00000166159), score: 0.66
LTV1LTV1 homolog (S. cerevisiae) (ENSG00000135521), score: -0.54
LZICleucine zipper and CTNNBIP1 domain containing (ENSG00000162441), score: -0.66
MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: -0.69
MARCH7membrane-associated ring finger (C3HC4) 7 (ENSG00000136536), score: -0.63
MATKmegakaryocyte-associated tyrosine kinase (ENSG00000007264), score: 0.7
MC4Rmelanocortin 4 receptor (ENSG00000166603), score: 0.61
MDM4Mdm4 p53 binding protein homolog (mouse) (ENSG00000198625), score: -0.63
MED7mediator complex subunit 7 (ENSG00000155868), score: -0.59
MGAT5Bmannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B (ENSG00000167889), score: 0.67
MKL1megakaryoblastic leukemia (translocation) 1 (ENSG00000196588), score: 0.72
MKL2MKL/myocardin-like 2 (ENSG00000186260), score: 0.62
MTPAPmitochondrial poly(A) polymerase (ENSG00000107951), score: -0.67
MYNNmyoneurin (ENSG00000085274), score: -0.55
NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (ENSG00000103174), score: 0.68
NCDNneurochondrin (ENSG00000020129), score: 0.65
NEDD1neural precursor cell expressed, developmentally down-regulated 1 (ENSG00000139350), score: -0.57
NETO1neuropilin (NRP) and tolloid (TLL)-like 1 (ENSG00000166342), score: 0.65
NHLRC2NHL repeat containing 2 (ENSG00000196865), score: -0.54
NTSR1neurotensin receptor 1 (high affinity) (ENSG00000101188), score: 0.65
OCA2oculocutaneous albinism II (ENSG00000104044), score: 0.72
OPN4opsin 4 (ENSG00000122375), score: 0.97
OPRL1opiate receptor-like 1 (ENSG00000125510), score: 0.67
PAN3PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (ENSG00000152520), score: -0.56
PCDH19protocadherin 19 (ENSG00000165194), score: 0.62
PCSK1proprotein convertase subtilisin/kexin type 1 (ENSG00000175426), score: 0.62
PDE8Bphosphodiesterase 8B (ENSG00000113231), score: 0.66
PDZD7PDZ domain containing 7 (ENSG00000186862), score: 0.64
PEX13peroxisomal biogenesis factor 13 (ENSG00000162928), score: -0.58
PIK3C2Aphosphoinositide-3-kinase, class 2, alpha polypeptide (ENSG00000011405), score: -0.55
PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (ENSG00000105647), score: 0.62
PLEKHM2pleckstrin homology domain containing, family M (with RUN domain) member 2 (ENSG00000116786), score: 0.64
POLR3Apolymerase (RNA) III (DNA directed) polypeptide A, 155kDa (ENSG00000148606), score: 0.65
PRPF19PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae) (ENSG00000110107), score: 0.62
PSDpleckstrin and Sec7 domain containing (ENSG00000059915), score: 0.66
PTHLHparathyroid hormone-like hormone (ENSG00000087494), score: 0.64
RAB40CRAB40C, member RAS oncogene family (ENSG00000197562), score: 0.79
RANGAP1Ran GTPase activating protein 1 (ENSG00000100401), score: 0.64
RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.68
RIMS3regulating synaptic membrane exocytosis 3 (ENSG00000117016), score: 0.62
ROCK1Rho-associated, coiled-coil containing protein kinase 1 (ENSG00000067900), score: -0.73
RSPO2R-spondin 2 homolog (Xenopus laevis) (ENSG00000147655), score: 0.62
RTN4RL1reticulon 4 receptor-like 1 (ENSG00000185924), score: 0.62
SARSseryl-tRNA synthetase (ENSG00000031698), score: 0.62
SCN3Bsodium channel, voltage-gated, type III, beta (ENSG00000166257), score: 0.62
SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: 0.65
SIN3BSIN3 homolog B, transcription regulator (yeast) (ENSG00000127511), score: 0.66
SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (ENSG00000160326), score: 0.66
SLC6A7solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 (ENSG00000011083), score: 0.63
SMAD5SMAD family member 5 (ENSG00000113658), score: -0.61
SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: -0.54
SPDEFSAM pointed domain containing ets transcription factor (ENSG00000124664), score: 0.75
SREBF2sterol regulatory element binding transcription factor 2 (ENSG00000198911), score: 0.62
SSTsomatostatin (ENSG00000157005), score: 0.66
ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (ENSG00000117069), score: 0.69
STOML1stomatin (EPB72)-like 1 (ENSG00000067221), score: 0.68
SYT17synaptotagmin XVII (ENSG00000103528), score: 0.69
TBC1D23TBC1 domain family, member 23 (ENSG00000036054), score: -0.63
TDRD3tudor domain containing 3 (ENSG00000083544), score: -0.63
TGFBR1transforming growth factor, beta receptor 1 (ENSG00000106799), score: -0.73
THOC2THO complex 2 (ENSG00000125676), score: -0.6
TMEM175transmembrane protein 175 (ENSG00000127419), score: 0.65
TMEM179transmembrane protein 179 (ENSG00000189203), score: 0.67
TP53INP1tumor protein p53 inducible nuclear protein 1 (ENSG00000164938), score: -0.55
TRPM2transient receptor potential cation channel, subfamily M, member 2 (ENSG00000142185), score: 0.63
TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: -0.67
UNC5Aunc-5 homolog A (C. elegans) (ENSG00000113763), score: 0.67
USP3ubiquitin specific peptidase 3 (ENSG00000140455), score: -0.55
VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: -0.61
WDR37WD repeat domain 37 (ENSG00000047056), score: 0.67
XRN15'-3' exoribonuclease 1 (ENSG00000114127), score: -0.58
XRN25'-3' exoribonuclease 2 (ENSG00000088930), score: -0.54
ZDHHC22zinc finger, DHHC-type containing 22 (ENSG00000177108), score: 0.68
ZER1zer-1 homolog (C. elegans) (ENSG00000160445), score: 0.65
ZFAND6zinc finger, AN1-type domain 6 (ENSG00000086666), score: -0.58
Non-Entrez genes
Unknown, score:
HELP
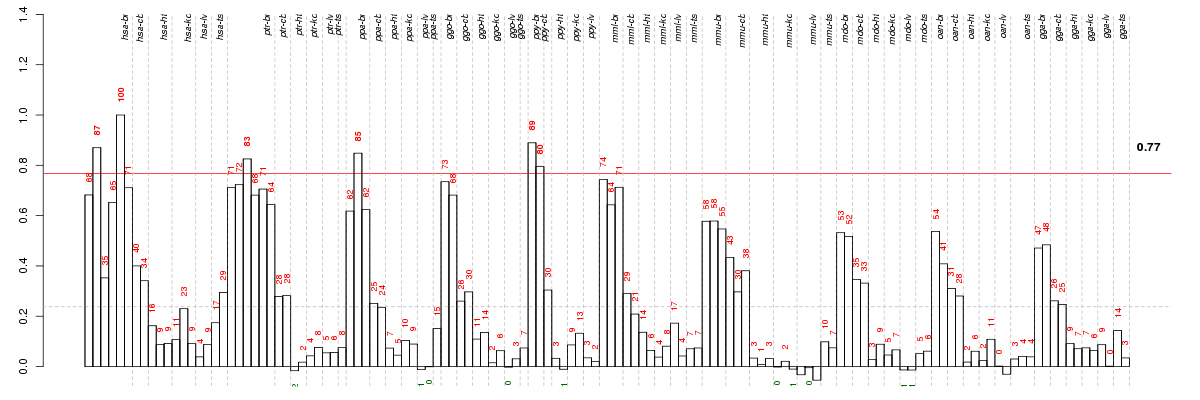
Conditions in the module, given in the same order as on the expression
plot above. Red color means over-expression, green under-expression in
the given condition.
The barplot below shows the condition (sample) scores. A separate bar
is shown for each sample, its height is the corresponding score of the
sample in the module. The red and green numbers on the bars are the
sample scores expressed in percents, i.e. 100% is 1.0.
The height of each bar corresponds to the weighted
mean expression of the module genes. The weights are the gene scores
of the module and they are positive for the genes listed in
red above and they are negative for
the genes that are listed in
green.
Bars going up correspond to samples listed in
red (the ones that are different
enough to included in the module). In these samples
the red module genes are highly
expressed, and the green
module genes are lowly expressed. The behavior of the genes is the
opposite for bars going down.
The red and green lines show the module thresholds, samples above
the red line and below the green line are included in the module.
The different species and tissues that were part of the study, are separated
by dashed vertical lines.
— Click on the Help button again to close this help window.
| Id | species | tissue | sex | individual |
| ppy_br_f_ca1 | ppy | br | f | _ |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| ppy_br_m_ca1 | ppy | br | m | _ |
| hsa_br_m7_ca1 | hsa | br | m | 7 |



© 2008-2010 Computational Biology Group, Department of Medical Genetics,
University of Lausanne, Switzerland