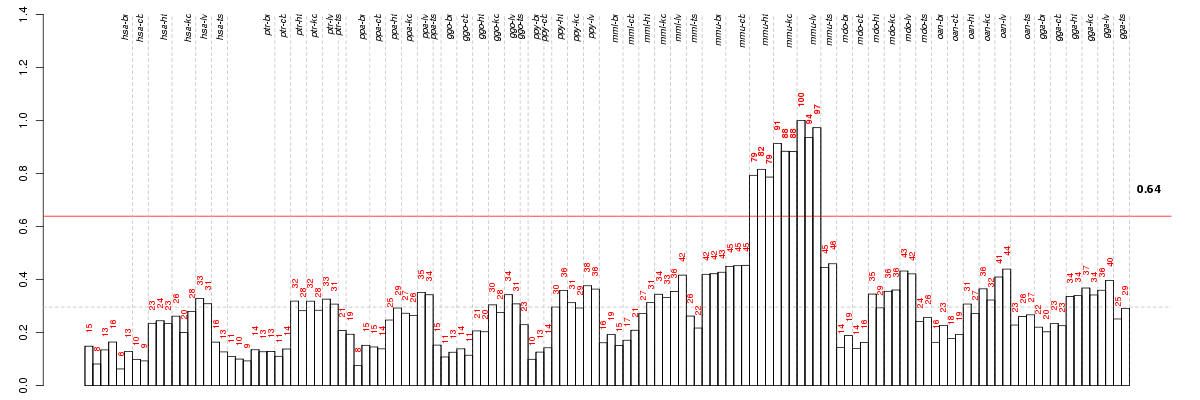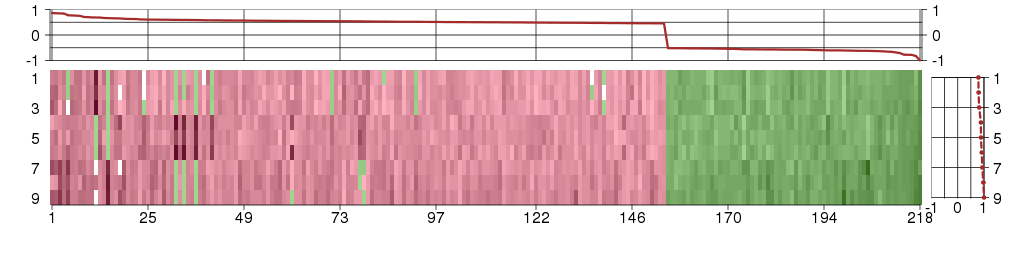

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The cellular synthesis of RNA on a template of DNA.
transcription from RNA polymerase III promoter
The synthesis of RNA from a DNA template by RNA polymerase III (Pol III), originating at a Pol III-specific promoter.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The cellular synthesis of RNA on a template of DNA.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.


A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.59 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.71 ABI2abl-interactor 2 (ENSG00000138443), score: -0.58 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.46 ADAP2ArfGAP with dual PH domains 2 (ENSG00000184060), score: 0.53 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (ENSG00000117448), score: 0.49 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.54 ANGEL1angel homolog 1 (Drosophila) (ENSG00000013523), score: -0.55 ANGEL2angel homolog 2 (Drosophila) (ENSG00000174606), score: 0.48 ANKRD16ankyrin repeat domain 16 (ENSG00000134461), score: 0.46 APBA3amyloid beta (A4) precursor protein-binding, family A, member 3 (ENSG00000011132), score: 0.51 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.6 ARFIP2ADP-ribosylation factor interacting protein 2 (ENSG00000132254), score: -0.53 ARHGAP42Rho GTPase activating protein 42 (ENSG00000165895), score: 0.53 ARMC7armadillo repeat containing 7 (ENSG00000125449), score: -0.56 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.6 ATP6V0A2ATPase, H+ transporting, lysosomal V0 subunit a2 (ENSG00000185344), score: 0.52 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: -0.53 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.83 BBS2Bardet-Biedl syndrome 2 (ENSG00000125124), score: -0.62 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: 0.56 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.61 C12orf72chromosome 12 open reading frame 72 (ENSG00000139160), score: 0.55 C16orf48chromosome 16 open reading frame 48 (ENSG00000124074), score: -0.57 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.63 C21orf63chromosome 21 open reading frame 63 (ENSG00000166979), score: -0.54 C2CD2C2 calcium-dependent domain containing 2 (ENSG00000157617), score: 0.47 C3orf37chromosome 3 open reading frame 37 (ENSG00000183624), score: -0.52 C3orf75chromosome 3 open reading frame 75 (ENSG00000163832), score: -0.57 C7orf23chromosome 7 open reading frame 23 (ENSG00000135185), score: 0.6 CASP8caspase 8, apoptosis-related cysteine peptidase (ENSG00000064012), score: 0.56 CCBL2cysteine conjugate-beta lyase 2 (ENSG00000137944), score: 0.57 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: 0.49 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.69 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.84 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.85 CGRRF1cell growth regulator with ring finger domain 1 (ENSG00000100532), score: 0.5 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.48 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: 0.49 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.58 CLSTN3calsyntenin 3 (ENSG00000139182), score: -0.52 CMTM6CKLF-like MARVEL transmembrane domain containing 6 (ENSG00000091317), score: 0.46 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.85 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.65 COMMD3COMM domain containing 3 (ENSG00000148444), score: 0.5 COPB2coatomer protein complex, subunit beta 2 (beta prime) (ENSG00000184432), score: 0.56 CPOXcoproporphyrinogen oxidase (ENSG00000080819), score: 0.53 CROTcarnitine O-octanoyltransferase (ENSG00000005469), score: 0.6 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: 0.52 CYBRD1cytochrome b reductase 1 (ENSG00000071967), score: -0.6 DARSaspartyl-tRNA synthetase (ENSG00000115866), score: 0.47 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.55 DCLRE1ADNA cross-link repair 1A (ENSG00000198924), score: 0.48 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: -0.63 DLSTdihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) (ENSG00000119689), score: 0.46 DNAJC3DnaJ (Hsp40) homolog, subfamily C, member 3 (ENSG00000102580), score: 0.52 DUSP14dual specificity phosphatase 14 (ENSG00000161326), score: -0.56 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (ENSG00000158711), score: 0.57 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.61 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: 0.49 ERBB2IPerbb2 interacting protein (ENSG00000112851), score: 0.5 ERN1endoplasmic reticulum to nucleus signaling 1 (ENSG00000178607), score: 0.5 ETFDHelectron-transferring-flavoprotein dehydrogenase (ENSG00000171503), score: 0.55 EXOC6exocyst complex component 6 (ENSG00000138190), score: 0.5 FABP2fatty acid binding protein 2, intestinal (ENSG00000145384), score: 0.66 FAM105Bfamily with sequence similarity 105, member B (ENSG00000154124), score: 0.53 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: -0.54 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: 0.5 FAM59Afamily with sequence similarity 59, member A (ENSG00000141441), score: 0.48 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: 0.55 FBXW8F-box and WD repeat domain containing 8 (ENSG00000174989), score: 0.54 FCHO2FCH domain only 2 (ENSG00000157107), score: 0.55 FEZ1fasciculation and elongation protein zeta 1 (zygin I) (ENSG00000149557), score: -0.57 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (ENSG00000141429), score: 0.52 GBA2glucosidase, beta (bile acid) 2 (ENSG00000070610), score: -0.64 GFM2G elongation factor, mitochondrial 2 (ENSG00000164347), score: 0.48 GGPS1geranylgeranyl diphosphate synthase 1 (ENSG00000152904), score: -0.57 GM2AGM2 ganglioside activator (ENSG00000196743), score: 0.47 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: 0.51 GNEglucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase (ENSG00000159921), score: 0.46 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.63 GRPEL2GrpE-like 2, mitochondrial (E. coli) (ENSG00000164284), score: 0.49 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: 0.5 H1FXH1 histone family, member X (ENSG00000184897), score: -1 H6PDhexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (ENSG00000049239), score: 0.55 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: 0.51 HDDC2HD domain containing 2 (ENSG00000111906), score: -0.6 HELBhelicase (DNA) B (ENSG00000127311), score: 0.57 HERPUD1homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 (ENSG00000051108), score: 0.51 HINFPhistone H4 transcription factor (ENSG00000172273), score: -0.58 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.5 HNRNPRheterogeneous nuclear ribonucleoprotein R (ENSG00000125944), score: -0.62 IFFO1intermediate filament family orphan 1 (ENSG00000010295), score: -0.65 IGHMBP2immunoglobulin mu binding protein 2 (ENSG00000132740), score: -0.57 ITM2Aintegral membrane protein 2A (ENSG00000078596), score: -0.53 ITM2Bintegral membrane protein 2B (ENSG00000136156), score: 0.5 IVNS1ABPinfluenza virus NS1A binding protein (ENSG00000116679), score: 0.54 KIAA0100KIAA0100 (ENSG00000007202), score: 0.57 KIAA2018KIAA2018 (ENSG00000176542), score: 0.52 KRT80keratin 80 (ENSG00000167767), score: 0.54 LACE1lactation elevated 1 (ENSG00000135537), score: 0.46 LAMP2lysosomal-associated membrane protein 2 (ENSG00000005893), score: 0.54 LARP4BLa ribonucleoprotein domain family, member 4B (ENSG00000107929), score: 0.56 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.66 LIFRleukemia inhibitory factor receptor alpha (ENSG00000113594), score: 0.64 LPGAT1lysophosphatidylglycerol acyltransferase 1 (ENSG00000123684), score: 0.5 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 0.69 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.67 MAP3K5mitogen-activated protein kinase kinase kinase 5 (ENSG00000197442), score: 0.47 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.69 MARCH6membrane-associated ring finger (C3HC4) 6 (ENSG00000145495), score: 0.54 MBIPMAP3K12 binding inhibitory protein 1 (ENSG00000151332), score: -0.62 MCCC1methylcrotonoyl-CoA carboxylase 1 (alpha) (ENSG00000078070), score: 0.56 MCEEmethylmalonyl CoA epimerase (ENSG00000124370), score: 0.46 MCM3APminichromosome maintenance complex component 3 associated protein (ENSG00000160294), score: -0.59 MCRS1microspherule protein 1 (ENSG00000187778), score: -0.57 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.77 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.59 MEP1Ameprin A, alpha (PABA peptide hydrolase) (ENSG00000112818), score: 0.59 MEP1Bmeprin A, beta (ENSG00000141434), score: 0.59 MFN1mitofusin 1 (ENSG00000171109), score: 0.53 MIA3melanoma inhibitory activity family, member 3 (ENSG00000154305), score: 0.52 MIPOL1mirror-image polydactyly 1 (ENSG00000151338), score: 0.59 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.55 MRASmuscle RAS oncogene homolog (ENSG00000158186), score: -0.53 MUTmethylmalonyl CoA mutase (ENSG00000146085), score: 0.48 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: -0.52 NDUFA6NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa (ENSG00000184983), score: 0.47 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.71 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.76 NHLRC2NHL repeat containing 2 (ENSG00000196865), score: 0.5 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.77 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.68 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: 0.49 OSGIN2oxidative stress induced growth inhibitor family member 2 (ENSG00000164823), score: -0.57 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.57 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.56 PAK1IP1PAK1 interacting protein 1 (ENSG00000111845), score: 0.54 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: -0.57 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.5 PARD3par-3 partitioning defective 3 homolog (C. elegans) (ENSG00000148498), score: 0.47 PCBD2pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 (ENSG00000132570), score: 0.46 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.55 PLAAphospholipase A2-activating protein (ENSG00000137055), score: 0.51 PLEKHN1pleckstrin homology domain containing, family N member 1 (ENSG00000187583), score: 0.51 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (ENSG00000113356), score: 0.46 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (ENSG00000161980), score: -0.53 PPM1Bprotein phosphatase, Mg2+/Mn2+ dependent, 1B (ENSG00000138032), score: 0.5 PPP1R7protein phosphatase 1, regulatory (inhibitor) subunit 7 (ENSG00000115685), score: -0.59 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha (ENSG00000066027), score: 0.58 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: -0.6 PRKAR1Bprotein kinase, cAMP-dependent, regulatory, type I, beta (ENSG00000188191), score: -0.54 PRKD3protein kinase D3 (ENSG00000115825), score: 0.6 PRLRprolactin receptor (ENSG00000113494), score: 0.52 PSMA4proteasome (prosome, macropain) subunit, alpha type, 4 (ENSG00000041357), score: 0.56 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (ENSG00000197170), score: 0.57 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (ENSG00000152229), score: 0.55 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: 0.58 RAB22ARAB22A, member RAS oncogene family (ENSG00000124209), score: 0.52 RALGAPA2Ral GTPase activating protein, alpha subunit 2 (catalytic) (ENSG00000188559), score: 0.54 RDM1RAD52 motif 1 (ENSG00000187456), score: 0.47 RDXradixin (ENSG00000137710), score: 0.54 RFESDRieske (Fe-S) domain containing (ENSG00000175449), score: 0.51 RNF139ring finger protein 139 (ENSG00000170881), score: 0.63 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.51 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.62 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: -0.56 SCCPDHsaccharopine dehydrogenase (putative) (ENSG00000143653), score: -0.61 SEP1515 kDa selenoprotein (ENSG00000183291), score: 0.52 SEPT6septin 6 (ENSG00000125354), score: -0.6 SERINC3serine incorporator 3 (ENSG00000132824), score: 0.6 SFMBT2Scm-like with four mbt domains 2 (ENSG00000198879), score: -0.64 SGPP1sphingosine-1-phosphate phosphatase 1 (ENSG00000126821), score: 0.46 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: -0.66 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.54 SLC16A10solute carrier family 16, member 10 (aromatic amino acid transporter) (ENSG00000112394), score: 0.54 SLC18A1solute carrier family 18 (vesicular monoamine), member 1 (ENSG00000036565), score: 0.58 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: 0.56 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: -0.57 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.48 SLC39A6solute carrier family 39 (zinc transporter), member 6 (ENSG00000141424), score: -0.63 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.46 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.78 SNAPC3small nuclear RNA activating complex, polypeptide 3, 50kDa (ENSG00000164975), score: -0.54 SNX11sorting nexin 11 (ENSG00000002919), score: -0.55 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.75 STAMBPSTAM binding protein (ENSG00000124356), score: -0.52 TBCELtubulin folding cofactor E-like (ENSG00000154114), score: 0.49 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: -0.61 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.57 TGDSTDP-glucose 4,6-dehydratase (ENSG00000088451), score: 0.51 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (ENSG00000135966), score: 0.49 TM2D2TM2 domain containing 2 (ENSG00000169490), score: 0.48 TM9SF2transmembrane 9 superfamily member 2 (ENSG00000125304), score: 0.47 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: 0.55 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.59 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.54 TMEM14Atransmembrane protein 14A (ENSG00000096092), score: -0.59 TMEM20transmembrane protein 20 (ENSG00000176273), score: 0.47 TMEM64transmembrane protein 64 (ENSG00000180694), score: 0.6 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.5 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (ENSG00000145779), score: 0.53 TUBE1tubulin, epsilon 1 (ENSG00000074935), score: -0.58 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: 0.58 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.7 UBTD2ubiquitin domain containing 2 (ENSG00000168246), score: -0.61 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.48 VPS45vacuolar protein sorting 45 homolog (S. cerevisiae) (ENSG00000136631), score: -0.53 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: 0.48 WDR36WD repeat domain 36 (ENSG00000134987), score: 0.59 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.77 WRBtryptophan rich basic protein (ENSG00000182093), score: -0.58 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.65 ZBTB49zinc finger and BTB domain containing 49 (ENSG00000168826), score: -0.52 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.86 ZC4H2zinc finger, C4H2 domain containing (ENSG00000126970), score: -0.53 ZMYND19zinc finger, MYND-type containing 19 (ENSG00000165724), score: -0.52 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.77
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |