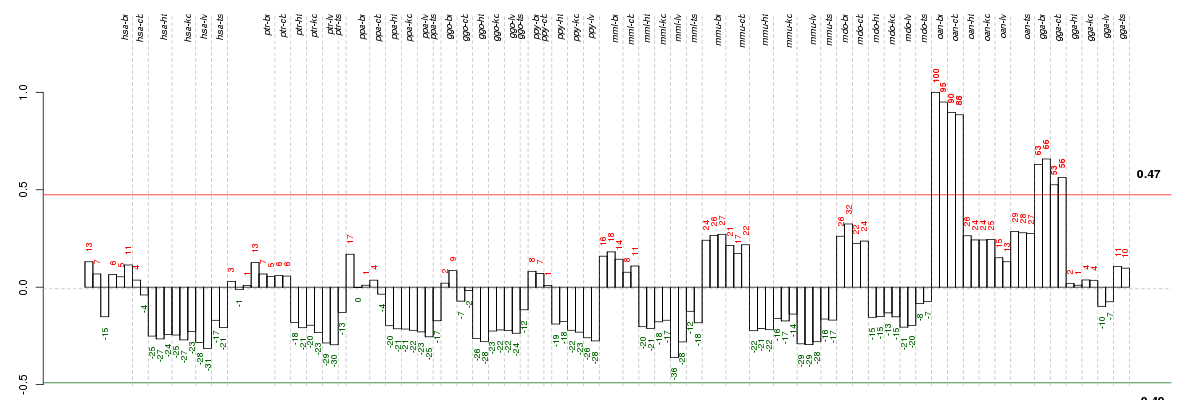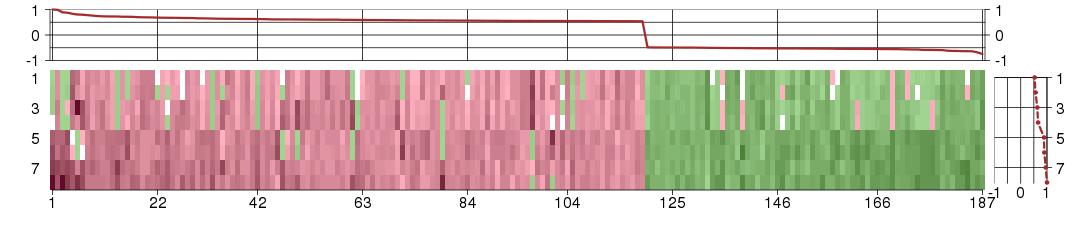

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
ligand-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
neurotransmitter binding
Interacting selectively and non-covalently with a neurotransmitter, any chemical substance that is capable of transmitting (or inhibiting the transmission of) a nerve impulse from a neuron to another cell.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.64 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: -0.53 ANKRA2ankyrin repeat, family A (RFXANK-like), 2 (ENSG00000164331), score: 0.64 ANKRD27ankyrin repeat domain 27 (VPS9 domain) (ENSG00000105186), score: -0.53 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: -0.51 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: -0.55 ARIH2ariadne homolog 2 (Drosophila) (ENSG00000177479), score: -0.55 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.6 ATRXalpha thalassemia/mental retardation syndrome X-linked (ENSG00000085224), score: 0.54 AURKAIP1aurora kinase A interacting protein 1 (ENSG00000175756), score: -0.52 AVPR1Barginine vasopressin receptor 1B (ENSG00000198049), score: 1 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.59 BCDIN3DBCDIN3 domain containing (ENSG00000186666), score: -0.53 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.6 BMP3bone morphogenetic protein 3 (ENSG00000152785), score: 0.68 C12orf29chromosome 12 open reading frame 29 (ENSG00000133641), score: 0.56 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.57 C14orf37chromosome 14 open reading frame 37 (ENSG00000139971), score: 0.73 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.53 C1orf25chromosome 1 open reading frame 25 (ENSG00000121486), score: 0.59 C20orf4chromosome 20 open reading frame 4 (ENSG00000131043), score: 0.55 C5orf24chromosome 5 open reading frame 24 (ENSG00000181904), score: 0.63 C6orf106chromosome 6 open reading frame 106 (ENSG00000196821), score: -0.63 C7orf27chromosome 7 open reading frame 27 (ENSG00000106009), score: -0.49 CACHD1cache domain containing 1 (ENSG00000158966), score: 0.55 CACNG2calcium channel, voltage-dependent, gamma subunit 2 (ENSG00000166862), score: 0.57 CASD1CAS1 domain containing 1 (ENSG00000127995), score: 0.58 CASTcalpastatin (ENSG00000153113), score: -0.53 CDC40cell division cycle 40 homolog (S. cerevisiae) (ENSG00000168438), score: 0.59 CDK17cyclin-dependent kinase 17 (ENSG00000059758), score: 0.61 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.51 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.63 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.54 CHRNB4cholinergic receptor, nicotinic, beta 4 (ENSG00000117971), score: 0.58 CLCN3chloride channel 3 (ENSG00000109572), score: 0.68 CLVS2clavesin 2 (ENSG00000146352), score: 0.58 CNGB1cyclic nucleotide gated channel beta 1 (ENSG00000070729), score: 0.58 CNTN5contactin 5 (ENSG00000149972), score: 0.99 COL25A1collagen, type XXV, alpha 1 (ENSG00000188517), score: 0.67 COMMD2COMM domain containing 2 (ENSG00000114744), score: 0.61 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -0.55 CPLX4complexin 4 (ENSG00000166569), score: 0.73 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.64 CTNND1catenin (cadherin-associated protein), delta 1 (ENSG00000198561), score: -0.63 CUL4Acullin 4A (ENSG00000139842), score: -0.75 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.54 DCNdecorin (ENSG00000011465), score: -0.51 DCTN4dynactin 4 (p62) (ENSG00000132912), score: 0.73 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.69 DUSP19dual specificity phosphatase 19 (ENSG00000162999), score: 0.6 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.55 ELP2elongation protein 2 homolog (S. cerevisiae) (ENSG00000134759), score: -0.54 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.55 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.73 EPHX4epoxide hydrolase 4 (ENSG00000172031), score: 0.55 FADDFas (TNFRSF6)-associated via death domain (ENSG00000168040), score: -0.48 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.52 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: 0.55 FAM164Afamily with sequence similarity 164, member A (ENSG00000104427), score: 0.63 FAM63Bfamily with sequence similarity 63, member B (ENSG00000128923), score: 0.54 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.63 FRMPD4FERM and PDZ domain containing 4 (ENSG00000169933), score: 0.57 FUT11fucosyltransferase 11 (alpha (1,3) fucosyltransferase) (ENSG00000196968), score: -0.55 GAAglucosidase, alpha; acid (ENSG00000171298), score: -0.62 GABRA2gamma-aminobutyric acid (GABA) A receptor, alpha 2 (ENSG00000151834), score: 0.67 GABRA4gamma-aminobutyric acid (GABA) A receptor, alpha 4 (ENSG00000109158), score: 0.69 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.61 GAD2glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) (ENSG00000136750), score: 0.57 GAMTguanidinoacetate N-methyltransferase (ENSG00000130005), score: -0.49 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 0.83 GPR149G protein-coupled receptor 149 (ENSG00000174948), score: 0.61 GPR85G protein-coupled receptor 85 (ENSG00000164604), score: 0.54 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.68 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.57 HEATR5BHEAT repeat containing 5B (ENSG00000008869), score: 0.62 HECW2HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 (ENSG00000138411), score: 0.56 HERPUD1homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 (ENSG00000051108), score: -0.56 HHLA1HERV-H LTR-associating 1 (ENSG00000132297), score: 0.87 HSF4heat shock transcription factor 4 (ENSG00000102878), score: -0.54 ICMTisoprenylcysteine carboxyl methyltransferase (ENSG00000116237), score: -0.52 IL12RB2interleukin 12 receptor, beta 2 (ENSG00000081985), score: 0.55 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.71 IL1RAPL2interleukin 1 receptor accessory protein-like 2 (ENSG00000189108), score: 0.8 INPPL1inositol polyphosphate phosphatase-like 1 (ENSG00000165458), score: -0.5 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: 0.74 KBTBD3kelch repeat and BTB (POZ) domain containing 3 (ENSG00000182359), score: 0.7 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.59 KIAA0368KIAA0368 (ENSG00000136813), score: -0.53 KIAA2022KIAA2022 (ENSG00000050030), score: 0.56 KLHDC2kelch domain containing 2 (ENSG00000165516), score: 0.55 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.64 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.58 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: -0.55 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.78 LOC100293817similar to hCG1811893 (ENSG00000144460), score: 0.55 LOC100293905similar to gastrin-releasing peptide receptor (ENSG00000126010), score: 0.72 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (ENSG00000126243), score: 0.61 LRP12low density lipoprotein receptor-related protein 12 (ENSG00000147650), score: 0.58 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.62 LRRC33leucine rich repeat containing 33 (ENSG00000174004), score: -0.5 LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.59 LSAMPlimbic system-associated membrane protein (ENSG00000185565), score: 0.56 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.51 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: -0.51 MAP3K3mitogen-activated protein kinase kinase kinase 3 (ENSG00000198909), score: -0.49 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: -0.53 MCF2MCF.2 cell line derived transforming sequence (ENSG00000101977), score: 0.61 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.69 MLXIPMLX interacting protein (ENSG00000175727), score: -0.63 MYO16myosin XVI (ENSG00000041515), score: 0.6 MYOGmyogenin (myogenic factor 4) (ENSG00000122180), score: 0.55 NARFLnuclear prelamin A recognition factor-like (ENSG00000103245), score: -0.5 NECAP2NECAP endocytosis associated 2 (ENSG00000157191), score: -0.62 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (ENSG00000109320), score: -0.56 NGBneuroglobin (ENSG00000165553), score: 0.57 NMNAT3nicotinamide nucleotide adenylyltransferase 3 (ENSG00000163864), score: -0.49 NPATnuclear protein, ataxia-telangiectasia locus (ENSG00000149308), score: 0.54 NPY2Rneuropeptide Y receptor Y2 (ENSG00000185149), score: 0.61 NTSneurotensin (ENSG00000133636), score: 0.64 OFCC1orofacial cleft 1 candidate 1 (ENSG00000181355), score: 0.57 OLIG3oligodendrocyte transcription factor 3 (ENSG00000177468), score: 0.8 PAFAH1B2platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) (ENSG00000168092), score: 0.55 PCGF3polycomb group ring finger 3 (ENSG00000185619), score: -0.53 PCGF5polycomb group ring finger 5 (ENSG00000180628), score: -0.54 PDE5Aphosphodiesterase 5A, cGMP-specific (ENSG00000138735), score: 0.57 PFKFB46-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (ENSG00000114268), score: 0.55 PNOCprepronociceptin (ENSG00000168081), score: 0.54 POLGpolymerase (DNA directed), gamma (ENSG00000140521), score: -0.54 PTP4A2protein tyrosine phosphatase type IVA, member 2 (ENSG00000184007), score: -0.55 PTPRAprotein tyrosine phosphatase, receptor type, A (ENSG00000132670), score: 0.56 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: -0.52 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.57 RAB3GAP2RAB3 GTPase activating protein subunit 2 (non-catalytic) (ENSG00000118873), score: 0.65 RECQL5RecQ protein-like 5 (ENSG00000108469), score: -0.51 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.55 RIC3resistance to inhibitors of cholinesterase 3 homolog (C. elegans) (ENSG00000166405), score: 0.57 RNF114ring finger protein 114 (ENSG00000124226), score: -0.55 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: 0.63 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: -0.59 RPS6KC1ribosomal protein S6 kinase, 52kDa, polypeptide 1 (ENSG00000136643), score: 0.65 RSPO3R-spondin 3 homolog (Xenopus laevis) (ENSG00000146374), score: 0.6 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (ENSG00000197632), score: 0.55 SH2B3SH2B adaptor protein 3 (ENSG00000111252), score: -0.52 SLC25A37solute carrier family 25, member 37 (ENSG00000147454), score: -0.52 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.57 SLC35A5solute carrier family 35, member A5 (ENSG00000138459), score: -0.54 SLC5A7solute carrier family 5 (choline transporter), member 7 (ENSG00000115665), score: 0.59 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.59 SNX25sorting nexin 25 (ENSG00000109762), score: 0.56 SNX5sorting nexin 5 (ENSG00000089006), score: -0.49 SPASTspastin (ENSG00000021574), score: 0.56 SPON1spondin 1, extracellular matrix protein (ENSG00000152268), score: 0.56 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (ENSG00000124214), score: 0.57 STK40serine/threonine kinase 40 (ENSG00000196182), score: -0.49 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.58 STX12syntaxin 12 (ENSG00000117758), score: 0.65 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.54 SYNJ1synaptojanin 1 (ENSG00000159082), score: 0.55 TAPT1transmembrane anterior posterior transformation 1 (ENSG00000169762), score: -0.52 TBC1D19TBC1 domain family, member 19 (ENSG00000109680), score: 0.55 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: 0.59 TDRD7tudor domain containing 7 (ENSG00000196116), score: -0.57 TFAP2Dtranscription factor AP-2 delta (activating enhancer binding protein 2 delta) (ENSG00000008197), score: 0.61 THOC7THO complex 7 homolog (Drosophila) (ENSG00000163634), score: -0.51 THSD7Athrombospondin, type I, domain containing 7A (ENSG00000005108), score: 0.67 TMEM117transmembrane protein 117 (ENSG00000139173), score: 0.76 TMEM22transmembrane protein 22 (ENSG00000168917), score: 0.54 TNMDtenomodulin (ENSG00000000005), score: 0.55 TOM1L1target of myb1 (chicken)-like 1 (ENSG00000141198), score: -0.53 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: -0.59 TPBGtrophoblast glycoprotein (ENSG00000146242), score: 0.67 TPRA1transmembrane protein, adipocyte asscociated 1 (ENSG00000163870), score: -0.49 TRDMT1tRNA aspartic acid methyltransferase 1 (ENSG00000107614), score: 0.55 TRPC1transient receptor potential cation channel, subfamily C, member 1 (ENSG00000144935), score: 0.6 TRPC5transient receptor potential cation channel, subfamily C, member 5 (ENSG00000072315), score: 0.67 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: -0.53 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.58 UBL3ubiquitin-like 3 (ENSG00000122042), score: 0.54 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.68 WACWW domain containing adaptor with coiled-coil (ENSG00000095787), score: -0.49 WDR26WD repeat domain 26 (ENSG00000162923), score: -0.54 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.62 WNT3Awingless-type MMTV integration site family, member 3A (ENSG00000154342), score: 0.89 WNT4wingless-type MMTV integration site family, member 4 (ENSG00000162552), score: 0.59 ZC3H12Bzinc finger CCCH-type containing 12B (ENSG00000102053), score: 0.55 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.72 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.49
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_cb_m_ca1 | gga | cb | m | _ |
| gga_cb_f_ca1 | gga | cb | f | _ |
| gga_br_m_ca1 | gga | br | m | _ |
| gga_br_f_ca1 | gga | br | f | _ |
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| oan_br_m_ca1 | oan | br | m | _ |