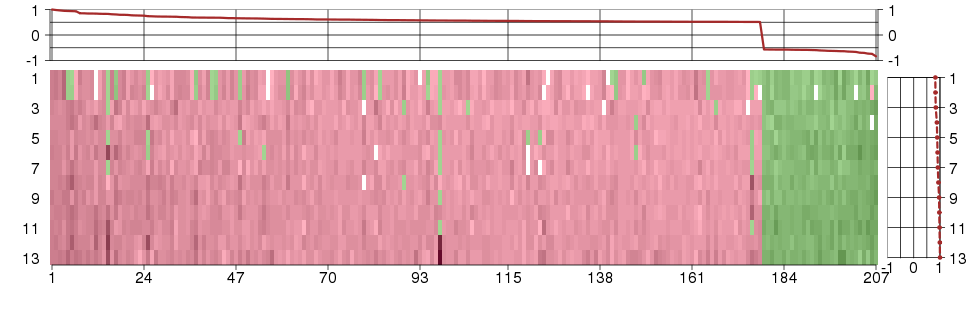

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
neurogenesis
Generation of cells within the nervous system.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
generation of neurons
The process by which nerve cells are generated. This includes the production of neuroblasts and their differentiation into neurons.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
neurogenesis
Generation of cells within the nervous system.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
potassium channel activity
Catalysis of facilitated diffusion of a potassium ion (by an energy-independent process) involving passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
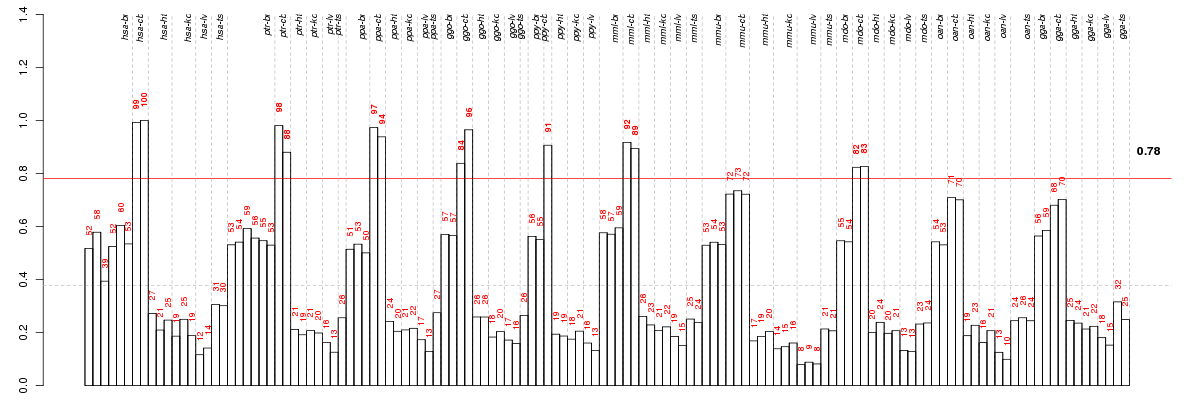
ABCC8ATP-binding cassette, sub-family C (CFTR/MRP), member 8 (ENSG00000006071), score: 0.6 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (ENSG00000172350), score: 0.51 ABLIM1actin binding LIM protein 1 (ENSG00000099204), score: 0.58 ADAM22ADAM metallopeptidase domain 22 (ENSG00000008277), score: 0.55 AKAP13A kinase (PRKA) anchor protein 13 (ENSG00000170776), score: -0.65 AP4B1adaptor-related protein complex 4, beta 1 subunit (ENSG00000134262), score: 0.53 ARPC2actin related protein 2/3 complex, subunit 2, 34kDa (ENSG00000163466), score: -0.58 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (ENSG00000170340), score: -0.63 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.6 BPTFbromodomain PHD finger transcription factor (ENSG00000171634), score: 0.59 BTBD3BTB (POZ) domain containing 3 (ENSG00000132640), score: 0.56 C13orf23chromosome 13 open reading frame 23 (ENSG00000120685), score: 0.52 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.7 C17orf28chromosome 17 open reading frame 28 (ENSG00000167861), score: 0.52 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: 0.54 C6orf115chromosome 6 open reading frame 115 (ENSG00000146386), score: -0.84 C7orf16chromosome 7 open reading frame 16 (ENSG00000106341), score: 0.6 C8orf79chromosome 8 open reading frame 79 (ENSG00000170941), score: 0.79 CA10carbonic anhydrase X (ENSG00000154975), score: 0.57 CA8carbonic anhydrase VIII (ENSG00000178538), score: 0.58 CACNA1Gcalcium channel, voltage-dependent, T type, alpha 1G subunit (ENSG00000006283), score: 0.56 CACNA1Icalcium channel, voltage-dependent, T type, alpha 1I subunit (ENSG00000100346), score: 0.51 CAMKK2calcium/calmodulin-dependent protein kinase kinase 2, beta (ENSG00000110931), score: 0.63 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.83 CCNJLcyclin J-like (ENSG00000135083), score: 0.72 CDH18cadherin 18, type 2 (ENSG00000145526), score: 0.57 CDH23cadherin-related 23 (ENSG00000107736), score: 0.57 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.77 CDONCdon homolog (mouse) (ENSG00000064309), score: 0.64 CDR2Lcerebellar degeneration-related protein 2-like (ENSG00000109089), score: 0.59 CERKLceramide kinase-like (ENSG00000188452), score: 0.57 CHGBchromogranin B (secretogranin 1) (ENSG00000089199), score: 0.61 CHN2chimerin (chimaerin) 2 (ENSG00000106069), score: 0.65 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.84 CLVS2clavesin 2 (ENSG00000146352), score: 0.61 CNKSR2connector enhancer of kinase suppressor of Ras 2 (ENSG00000149970), score: 0.52 CNOT2CCR4-NOT transcription complex, subunit 2 (ENSG00000111596), score: 0.73 CNTN6contactin 6 (ENSG00000134115), score: 0.73 CNTNAP1contactin associated protein 1 (ENSG00000108797), score: 0.53 CPPED1calcineurin-like phosphoesterase domain containing 1 (ENSG00000103381), score: -0.61 CRAMP1LCrm, cramped-like (Drosophila) (ENSG00000007545), score: 0.51 CREBBPCREB binding protein (ENSG00000005339), score: 0.52 CRHR1corticotropin releasing hormone receptor 1 (ENSG00000120088), score: 0.6 CRTAMcytotoxic and regulatory T cell molecule (ENSG00000109943), score: 0.94 DHRS13dehydrogenase/reductase (SDR family) member 13 (ENSG00000167536), score: 0.65 DIAPH2diaphanous homolog 2 (Drosophila) (ENSG00000147202), score: -0.58 DNM3dynamin 3 (ENSG00000197959), score: 0.55 E4F1E4F transcription factor 1 (ENSG00000167967), score: 0.52 EBF1early B-cell factor 1 (ENSG00000164330), score: 0.59 ECE2endothelin converting enzyme 2 (ENSG00000145194), score: 0.58 EDC4enhancer of mRNA decapping 4 (ENSG00000038358), score: 0.52 EPC1enhancer of polycomb homolog 1 (Drosophila) (ENSG00000120616), score: 0.65 ERBB2IPerbb2 interacting protein (ENSG00000112851), score: -0.62 ESPNLespin-like (ENSG00000144488), score: 0.51 ETV6ets variant 6 (ENSG00000139083), score: -0.74 FAM131Bfamily with sequence similarity 131, member B (ENSG00000159784), score: 0.54 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.98 FGF3fibroblast growth factor 3 (ENSG00000186895), score: 0.57 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.95 FGF9fibroblast growth factor 9 (glia-activating factor) (ENSG00000102678), score: 0.51 FGFR1fibroblast growth factor receptor 1 (ENSG00000077782), score: 0.58 FKBP5FK506 binding protein 5 (ENSG00000096060), score: -0.73 FLT3fms-related tyrosine kinase 3 (ENSG00000122025), score: 0.63 FYCO1FYVE and coiled-coil domain containing 1 (ENSG00000163820), score: -0.57 FZD7frizzled homolog 7 (Drosophila) (ENSG00000155760), score: 0.72 GABRB2gamma-aminobutyric acid (GABA) A receptor, beta 2 (ENSG00000145864), score: 0.52 GALNT7UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (ENSG00000109586), score: 0.65 GBX2gastrulation brain homeobox 2 (ENSG00000168505), score: 0.54 GDNFglial cell derived neurotrophic factor (ENSG00000168621), score: 0.54 GHITMgrowth hormone inducible transmembrane protein (ENSG00000165678), score: -0.63 GIT2G protein-coupled receptor kinase interacting ArfGAP 2 (ENSG00000139436), score: 0.55 GLCEglucuronic acid epimerase (ENSG00000138604), score: 0.68 GLRA2glycine receptor, alpha 2 (ENSG00000101958), score: 0.56 GNG13guanine nucleotide binding protein (G protein), gamma 13 (ENSG00000127588), score: 0.56 GPR176G protein-coupled receptor 176 (ENSG00000166073), score: 0.54 GRB14growth factor receptor-bound protein 14 (ENSG00000115290), score: -0.57 GRIA4glutamate receptor, ionotrophic, AMPA 4 (ENSG00000152578), score: 0.62 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.68 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.62 HINFPhistone H4 transcription factor (ENSG00000172273), score: 0.58 IFFO2intermediate filament family orphan 2 (ENSG00000169991), score: 0.51 IGDCC3immunoglobulin superfamily, DCC subclass, member 3 (ENSG00000174498), score: 0.54 IGSF21immunoglobin superfamily, member 21 (ENSG00000117154), score: 0.52 JARID2jumonji, AT rich interactive domain 2 (ENSG00000008083), score: 0.52 JMJD1Cjumonji domain containing 1C (ENSG00000171988), score: 0.57 KCNA1potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) (ENSG00000111262), score: 0.56 KCNC1potassium voltage-gated channel, Shaw-related subfamily, member 1 (ENSG00000129159), score: 0.66 KCNK10potassium channel, subfamily K, member 10 (ENSG00000100433), score: 0.76 KCNK9potassium channel, subfamily K, member 9 (ENSG00000169427), score: 0.79 KCNQ2potassium voltage-gated channel, KQT-like subfamily, member 2 (ENSG00000075043), score: 0.54 KCNRGpotassium channel regulator (ENSG00000198553), score: 0.64 KCNT1potassium channel, subfamily T, member 1 (ENSG00000107147), score: 0.56 KCTD2potassium channel tetramerisation domain containing 2 (ENSG00000180901), score: 0.69 KIAA0240KIAA0240 (ENSG00000112624), score: 0.54 KIAA0922KIAA0922 (ENSG00000121210), score: -0.7 KIAA1217KIAA1217 (ENSG00000120549), score: -0.7 KIAA1409KIAA1409 (ENSG00000133958), score: 0.52 LBHlimb bud and heart development homolog (mouse) (ENSG00000213626), score: -0.58 LDB2LIM domain binding 2 (ENSG00000169744), score: -0.66 LDLRAP1low density lipoprotein receptor adaptor protein 1 (ENSG00000157978), score: 0.68 LOC100131509similar to inward rectifying K+ channel negative regulator Kir2.2v (ENSG00000184185), score: 0.56 LOC100294337hypothetical protein LOC100294337 (ENSG00000120071), score: 0.54 LRCH1leucine-rich repeats and calponin homology (CH) domain containing 1 (ENSG00000136141), score: 0.72 LRRC38leucine rich repeat containing 38 (ENSG00000162494), score: 0.62 LRRC3Bleucine rich repeat containing 3B (ENSG00000179796), score: 0.53 LYRM5LYR motif containing 5 (ENSG00000205707), score: -0.57 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.96 MAML1mastermind-like 1 (Drosophila) (ENSG00000161021), score: 0.53 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.79 MAN2A1mannosidase, alpha, class 2A, member 1 (ENSG00000112893), score: -0.57 MCF2LMCF.2 cell line derived transforming sequence-like (ENSG00000126217), score: 0.57 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.93 MED13Lmediator complex subunit 13-like (ENSG00000123066), score: 0.54 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.65 MLLmyeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) (ENSG00000118058), score: 0.62 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: -0.64 MSX2msh homeobox 2 (ENSG00000120149), score: 0.59 MTAPmethylthioadenosine phosphorylase (ENSG00000099810), score: -0.57 MTSS1Lmetastasis suppressor 1-like (ENSG00000132613), score: 0.53 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.84 NDPNorrie disease (pseudoglioma) (ENSG00000124479), score: 0.54 NFKBIDnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta (ENSG00000167604), score: 0.57 NIPAL3NIPA-like domain containing 3 (ENSG00000001461), score: 0.51 NKX6-3NK6 homeobox 3 (ENSG00000165066), score: 0.94 NMNAT2nicotinamide nucleotide adenylyltransferase 2 (ENSG00000157064), score: 0.53 NRG2neuregulin 2 (ENSG00000158458), score: 0.66 NRP1neuropilin 1 (ENSG00000099250), score: -0.66 NTF3neurotrophin 3 (ENSG00000185652), score: 0.53 NTMneurotrimin (ENSG00000182667), score: 0.51 ORAI2ORAI calcium release-activated calcium modulator 2 (ENSG00000160991), score: 0.54 PAG1phosphoprotein associated with glycosphingolipid microdomains 1 (ENSG00000076641), score: 0.52 PARP12poly (ADP-ribose) polymerase family, member 12 (ENSG00000059378), score: -0.59 PAX3paired box 3 (ENSG00000135903), score: 0.72 PAX6paired box 6 (ENSG00000007372), score: 0.84 PAXIP1PAX interacting (with transcription-activation domain) protein 1 (ENSG00000157212), score: 0.6 PDZD7PDZ domain containing 7 (ENSG00000186862), score: 0.68 PFKFB26-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (ENSG00000123836), score: -0.57 PHIPpleckstrin homology domain interacting protein (ENSG00000146247), score: 0.53 PIK3R3phosphoinositide-3-kinase, regulatory subunit 3 (gamma) (ENSG00000117461), score: 0.52 PLCB4phospholipase C, beta 4 (ENSG00000101333), score: 0.63 PLCXD3phosphatidylinositol-specific phospholipase C, X domain containing 3 (ENSG00000182836), score: 0.57 PLD5phospholipase D family, member 5 (ENSG00000180287), score: 0.58 PLK2polo-like kinase 2 (ENSG00000145632), score: -0.58 PPM1Hprotein phosphatase, Mg2+/Mn2+ dependent, 1H (ENSG00000111110), score: 0.55 PTCH1patched 1 (ENSG00000185920), score: 0.68 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.72 PTPRRprotein tyrosine phosphatase, receptor type, R (ENSG00000153233), score: 0.54 PVALBparvalbumin (ENSG00000100362), score: 0.59 PVRL3poliovirus receptor-related 3 (ENSG00000177707), score: -0.62 RAB26RAB26, member RAS oncogene family (ENSG00000167964), score: 0.54 RADILRas association and DIL domains (ENSG00000157927), score: 0.52 RASGEF1CRasGEF domain family, member 1C (ENSG00000146090), score: 0.61 RBM9RNA binding motif protein 9 (ENSG00000100320), score: 0.53 RESTRE1-silencing transcription factor (ENSG00000084093), score: -0.58 RIT2Ras-like without CAAX 2 (ENSG00000152214), score: 0.61 RMND5Arequired for meiotic nuclear division 5 homolog A (S. cerevisiae) (ENSG00000153561), score: 0.54 RNF122ring finger protein 122 (ENSG00000133874), score: 0.54 RNF144Aring finger protein 144A (ENSG00000151692), score: 0.56 RUNX1T1runt-related transcription factor 1; translocated to, 1 (cyclin D-related) (ENSG00000079102), score: 0.58 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: 0.53 SCRN1secernin 1 (ENSG00000136193), score: 0.52 SEL1L3sel-1 suppressor of lin-12-like 3 (C. elegans) (ENSG00000091490), score: 0.61 SETD5SET domain containing 5 (ENSG00000168137), score: 0.62 SHFSrc homology 2 domain containing F (ENSG00000138606), score: 0.66 SKAP2src kinase associated phosphoprotein 2 (ENSG00000005020), score: -0.57 SKIv-ski sarcoma viral oncogene homolog (avian) (ENSG00000157933), score: 0.68 SKOR1SKI family transcriptional corepressor 1 (ENSG00000188779), score: 0.81 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.85 SLC9A5solute carrier family 9 (sodium/hydrogen exchanger), member 5 (ENSG00000135740), score: 0.6 SLITRK4SLIT and NTRK-like family, member 4 (ENSG00000179542), score: 0.57 SNAP25synaptosomal-associated protein, 25kDa (ENSG00000132639), score: 0.53 SNED1sushi, nidogen and EGF-like domains 1 (ENSG00000162804), score: 0.62 SPHKAPSPHK1 interactor, AKAP domain containing (ENSG00000153820), score: 0.6 SPINK6serine peptidase inhibitor, Kazal type 6 (ENSG00000178172), score: 0.83 SPTBN5spectrin, beta, non-erythrocytic 5 (ENSG00000137877), score: 0.81 SRCIN1SRC kinase signaling inhibitor 1 (ENSG00000017373), score: 0.52 SRRM4serine/arginine repetitive matrix 4 (ENSG00000139767), score: 0.58 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.55 SYBUsyntabulin (syntaxin-interacting) (ENSG00000147642), score: 0.52 SYNPRsynaptoporin (ENSG00000163630), score: 0.54 SYT12synaptotagmin XII (ENSG00000173227), score: 0.56 SYT4synaptotagmin IV (ENSG00000132872), score: 0.63 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (ENSG00000109436), score: 0.52 TFAP2Etranscription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) (ENSG00000116819), score: 0.66 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.83 TLL1tolloid-like 1 (ENSG00000038295), score: 0.76 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: -0.59 TMEM63Ctransmembrane protein 63C (ENSG00000165548), score: 0.55 TNRC6Atrinucleotide repeat containing 6A (ENSG00000090905), score: 0.55 TP73tumor protein p73 (ENSG00000078900), score: 0.68 TRHDEthyrotropin-releasing hormone degrading enzyme (ENSG00000072657), score: 0.52 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.77 UNC5Bunc-5 homolog B (C. elegans) (ENSG00000107731), score: 0.55 UNC80unc-80 homolog (C. elegans) (ENSG00000144406), score: 0.54 UPF0639UPF0639 protein (ENSG00000175985), score: 0.67 VAT1Lvesicle amine transport protein 1 homolog (T. californica)-like (ENSG00000171724), score: 0.61 VSX1visual system homeobox 1 (ENSG00000100987), score: 0.74 WACWW domain containing adaptor with coiled-coil (ENSG00000095787), score: 0.63 WASF3WAS protein family, member 3 (ENSG00000132970), score: 0.53 WSCD1WSC domain containing 1 (ENSG00000179314), score: 0.55 YPEL4yippee-like 4 (Drosophila) (ENSG00000166793), score: 0.54 ZFHX3zinc finger homeobox 3 (ENSG00000140836), score: -0.61 ZFPM2zinc finger protein, multitype 2 (ENSG00000169946), score: 0.7 ZIC4Zic family member 4 (ENSG00000174963), score: 1 ZNF238zinc finger protein 238 (ENSG00000179456), score: 0.71 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.85 ZNF536zinc finger protein 536 (ENSG00000198597), score: 0.56
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| ptr_cb_f_ca1 | ptr | cb | f | _ |
| mml_cb_f_ca1 | mml | cb | f | _ |
| ppy_cb_f_ca1 | ppy | cb | f | _ |
| mml_cb_m_ca1 | mml | cb | m | _ |
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |
| hsa_cb_m_ca1 | hsa | cb | m | _ |
| hsa_cb_f_ca1 | hsa | cb | f | _ |