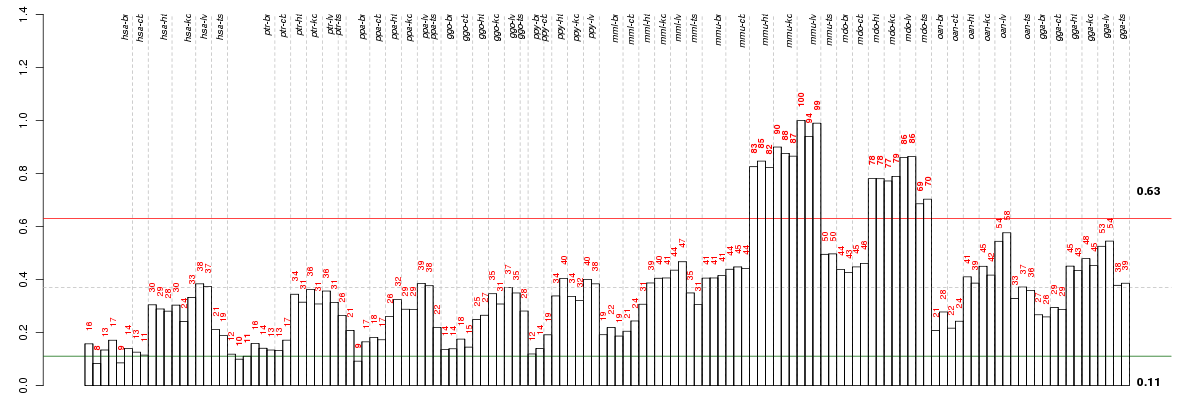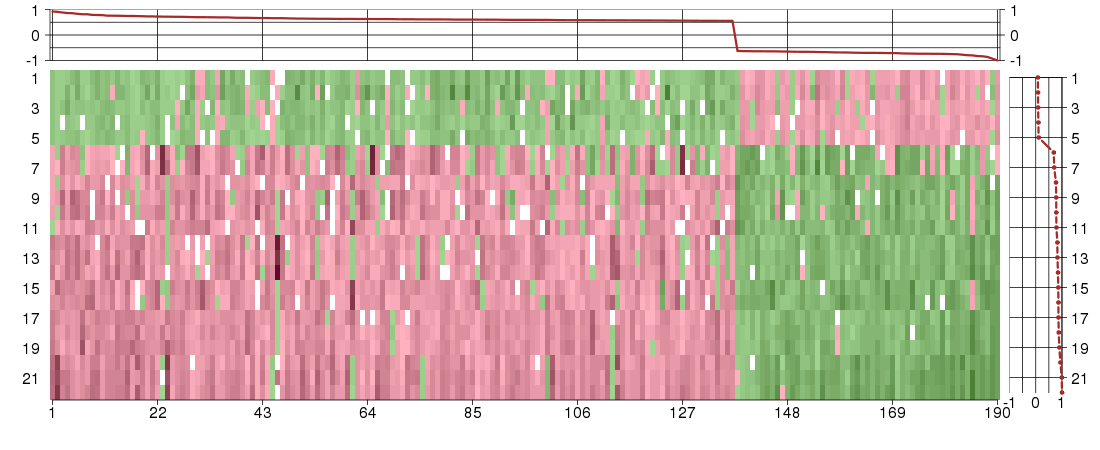

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
peroxisome
A small, membrane-bounded organelle that uses dioxygen (O2) to oxidize organic molecules; contains some enzymes that produce and others that degrade hydrogen peroxide (H2O2).
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
preribosome
Any complex of pre-rRNAs, ribosomal proteins, and associated proteins formed during ribosome biogenesis.
organelle membrane
The lipid bilayer surrounding an organelle.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to organelle membrane
Penetrating at least one phospholipid bilayer of an organelle membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
small-subunit processome
A large ribonucleoprotein complex that is an early preribosomal complex. In S. cerevisiae, it has a size of 80S and consists of the 35S pre-rRNA, early-associating ribosomal proteins most of which are part of the small ribosomal subunit, the U3 snoRNA and associated proteins.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
integral to organelle membrane
Penetrating at least one phospholipid bilayer of an organelle membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.75 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: 0.61 ABI2abl-interactor 2 (ENSG00000138443), score: -0.71 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.81 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: 0.57 ADAP2ArfGAP with dual PH domains 2 (ENSG00000184060), score: 0.6 AGPHD1aminoglycoside phosphotransferase domain containing 1 (ENSG00000188266), score: 0.56 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (ENSG00000117448), score: 0.58 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (ENSG00000119711), score: 0.58 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.67 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.67 AP2B1adaptor-related protein complex 2, beta 1 subunit (ENSG00000006125), score: -0.68 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 0.63 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.7 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.71 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.56 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: -0.75 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: 0.59 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.58 BRP44brain protein 44 (ENSG00000143158), score: 0.57 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.69 C17orf28chromosome 17 open reading frame 28 (ENSG00000167861), score: -0.67 C17orf39chromosome 17 open reading frame 39 (ENSG00000141034), score: 0.61 C1orf128chromosome 1 open reading frame 128 (ENSG00000057757), score: -0.73 C1orf83chromosome 1 open reading frame 83 (ENSG00000116205), score: 0.56 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.62 C3orf23chromosome 3 open reading frame 23 (ENSG00000179152), score: 0.63 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.74 CCBL2cysteine conjugate-beta lyase 2 (ENSG00000137944), score: 0.63 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: 0.59 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.83 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.92 CDK10cyclin-dependent kinase 10 (ENSG00000185324), score: -0.63 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.74 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.57 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.78 CLSTN3calsyntenin 3 (ENSG00000139182), score: -0.79 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.91 COPB2coatomer protein complex, subunit beta 2 (beta prime) (ENSG00000184432), score: 0.59 CROTcarnitine O-octanoyltransferase (ENSG00000005469), score: 0.63 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (ENSG00000070190), score: 0.67 DARSaspartyl-tRNA synthetase (ENSG00000115866), score: 0.6 DCTDdCMP deaminase (ENSG00000129187), score: -0.7 DDX41DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (ENSG00000183258), score: -0.67 DERL1Der1-like domain family, member 1 (ENSG00000136986), score: 0.56 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: -0.83 DLSTdihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) (ENSG00000119689), score: 0.56 DTNBP1dystrobrevin binding protein 1 (ENSG00000047579), score: 0.69 ECDecdysoneless homolog (Drosophila) (ENSG00000122882), score: 0.63 EEA1early endosome antigen 1 (ENSG00000102189), score: 0.72 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.75 ERN1endoplasmic reticulum to nucleus signaling 1 (ENSG00000178607), score: 0.58 ETFDHelectron-transferring-flavoprotein dehydrogenase (ENSG00000171503), score: 0.68 EVI5ecotropic viral integration site 5 (ENSG00000067208), score: 0.61 EXOC3exocyst complex component 3 (ENSG00000180104), score: -0.66 FAM160A1family with sequence similarity 160, member A1 (ENSG00000164142), score: 0.61 FAM173Bfamily with sequence similarity 173, member B (ENSG00000150756), score: 0.58 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: 0.57 FAM199Xfamily with sequence similarity 199, X-linked (ENSG00000123575), score: -0.7 FAM20Bfamily with sequence similarity 20, member B (ENSG00000116199), score: 0.66 FBXO2F-box protein 2 (ENSG00000116661), score: -0.72 FBXO22F-box protein 22 (ENSG00000167196), score: 0.56 FBXO3F-box protein 3 (ENSG00000110429), score: 0.88 FCHO2FCH domain only 2 (ENSG00000157107), score: 0.64 FEZ1fasciculation and elongation protein zeta 1 (zygin I) (ENSG00000149557), score: -0.75 FN3KRPfructosamine 3 kinase related protein (ENSG00000141560), score: -0.64 GALNTL1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1 (ENSG00000100626), score: -0.64 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.61 GGPS1geranylgeranyl diphosphate synthase 1 (ENSG00000152904), score: -0.64 GPAMglycerol-3-phosphate acyltransferase, mitochondrial (ENSG00000119927), score: 0.56 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.57 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: 0.68 H1FXH1 histone family, member X (ENSG00000184897), score: -0.8 HDDC2HD domain containing 2 (ENSG00000111906), score: -0.66 HDGFRP3hepatoma-derived growth factor, related protein 3 (ENSG00000166503), score: -0.64 HERPUD1homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 (ENSG00000051108), score: 0.6 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.6 HNRNPRheterogeneous nuclear ribonucleoprotein R (ENSG00000125944), score: -0.77 IDEinsulin-degrading enzyme (ENSG00000119912), score: 0.61 IL6STinterleukin 6 signal transducer (gp130, oncostatin M receptor) (ENSG00000134352), score: 0.56 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.62 IVNS1ABPinfluenza virus NS1A binding protein (ENSG00000116679), score: 0.57 KBTBD4kelch repeat and BTB (POZ) domain containing 4 (ENSG00000123444), score: -0.64 KCTD5potassium channel tetramerisation domain containing 5 (ENSG00000167977), score: -0.69 KIAA0174KIAA0174 (ENSG00000182149), score: -0.63 KIAA2018KIAA2018 (ENSG00000176542), score: 0.58 KLHL24kelch-like 24 (Drosophila) (ENSG00000114796), score: 0.59 KSR1kinase suppressor of ras 1 (ENSG00000141068), score: -0.68 LACE1lactation elevated 1 (ENSG00000135537), score: 0.64 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.76 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: 0.56 LIFRleukemia inhibitory factor receptor alpha (ENSG00000113594), score: 0.59 LMOD3leiomodin 3 (fetal) (ENSG00000163380), score: 0.58 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: 0.63 LPIN1lipin 1 (ENSG00000134324), score: 0.62 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 0.66 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.74 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: 0.6 MCCC1methylcrotonoyl-CoA carboxylase 1 (alpha) (ENSG00000078070), score: 0.57 MCEEmethylmalonyl CoA epimerase (ENSG00000124370), score: 0.63 MED1mediator complex subunit 1 (ENSG00000125686), score: 0.63 MFN1mitofusin 1 (ENSG00000171109), score: 0.83 MGST2microsomal glutathione S-transferase 2 (ENSG00000085871), score: -0.7 MIA3melanoma inhibitory activity family, member 3 (ENSG00000154305), score: 0.63 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.59 MPP6membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (ENSG00000105926), score: 0.59 MRASmuscle RAS oncogene homolog (ENSG00000158186), score: -0.74 MRPL13mitochondrial ribosomal protein L13 (ENSG00000172172), score: 0.62 MYH10myosin, heavy chain 10, non-muscle (ENSG00000133026), score: -0.73 NAA35N(alpha)-acetyltransferase 35, NatC auxiliary subunit (ENSG00000135040), score: 0.59 NAPEPLDN-acyl phosphatidylethanolamine phospholipase D (ENSG00000161048), score: -0.7 NDUFA6NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa (ENSG00000184983), score: 0.85 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -1 NMT2N-myristoyltransferase 2 (ENSG00000152465), score: -0.63 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.74 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.71 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.81 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.59 OSGIN2oxidative stress induced growth inhibitor family member 2 (ENSG00000164823), score: -0.65 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.75 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.68 PAPSS23'-phosphoadenosine 5'-phosphosulfate synthase 2 (ENSG00000198682), score: 0.61 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.62 PCBD2pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 (ENSG00000132570), score: 0.58 PCYT1Aphosphate cytidylyltransferase 1, choline, alpha (ENSG00000161217), score: 0.63 PDP2pyruvate dehyrogenase phosphatase catalytic subunit 2 (ENSG00000172840), score: 0.7 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.69 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.76 PHF5APHD finger protein 5A (ENSG00000100410), score: 0.63 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.64 PPIP5K2diphosphoinositol pentakisphosphate kinase 2 (ENSG00000145725), score: 0.62 PPP1R7protein phosphatase 1, regulatory (inhibitor) subunit 7 (ENSG00000115685), score: -0.65 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha (ENSG00000066027), score: 0.73 PRKD3protein kinase D3 (ENSG00000115825), score: 0.64 PRLRprolactin receptor (ENSG00000113494), score: 0.64 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.72 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: 0.56 PUS10pseudouridylate synthase 10 (ENSG00000162927), score: 0.61 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.7 QSER1glutamine and serine rich 1 (ENSG00000060749), score: 0.67 RASL11ARAS-like, family 11, member A (ENSG00000122035), score: -0.68 RC3H1ring finger and CCCH-type zinc finger domains 1 (ENSG00000135870), score: 0.58 RER1RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae) (ENSG00000157916), score: 0.57 RNF139ring finger protein 139 (ENSG00000170881), score: 0.69 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.74 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.71 RTN4IP1reticulon 4 interacting protein 1 (ENSG00000130347), score: 0.6 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.7 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.65 SEPSECSSep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (ENSG00000109618), score: 0.65 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.56 SLC16A10solute carrier family 16, member 10 (aromatic amino acid transporter) (ENSG00000112394), score: 0.79 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: 0.71 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.55 SLC39A6solute carrier family 39 (zinc transporter), member 6 (ENSG00000141424), score: -0.67 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.72 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.93 SNRKSNF related kinase (ENSG00000163788), score: 0.57 SNX13sorting nexin 13 (ENSG00000071189), score: 0.7 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: 0.65 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.6 SYDE2synapse defective 1, Rho GTPase, homolog 2 (C. elegans) (ENSG00000097096), score: 0.59 TBCELtubulin folding cofactor E-like (ENSG00000154114), score: 0.58 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: -0.7 TCP11L2t-complex 11 (mouse)-like 2 (ENSG00000166046), score: 0.66 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.6 TERTtelomerase reverse transcriptase (ENSG00000164362), score: 0.56 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.71 TMCO4transmembrane and coiled-coil domains 4 (ENSG00000162542), score: 0.58 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.75 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.63 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.6 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.64 TSR1TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) (ENSG00000167721), score: 0.67 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.86 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.86 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.78 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: 0.57 UTP11LUTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) (ENSG00000183520), score: 0.6 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.73 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: 0.69 VPS45vacuolar protein sorting 45 homolog (S. cerevisiae) (ENSG00000136631), score: -0.66 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: 0.64 WDR36WD repeat domain 36 (ENSG00000134987), score: 0.58 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.76 WRBtryptophan rich basic protein (ENSG00000182093), score: -0.73 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.58 ZC4H2zinc finger, C4H2 domain containing (ENSG00000126970), score: -0.73 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.72 ZNHIT3zinc finger, HIT type 3 (ENSG00000108278), score: -0.66
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| ptr_br_m2_ca1 | ptr | br | m | 2 |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |