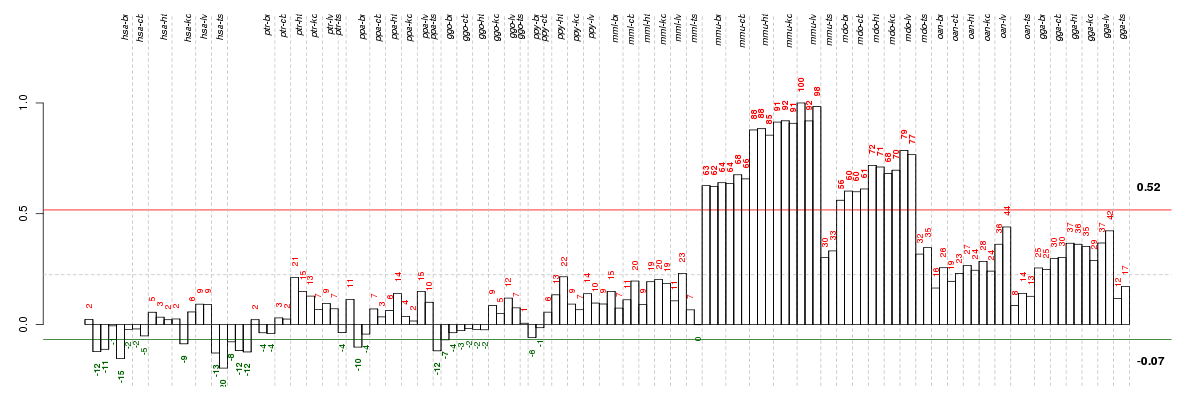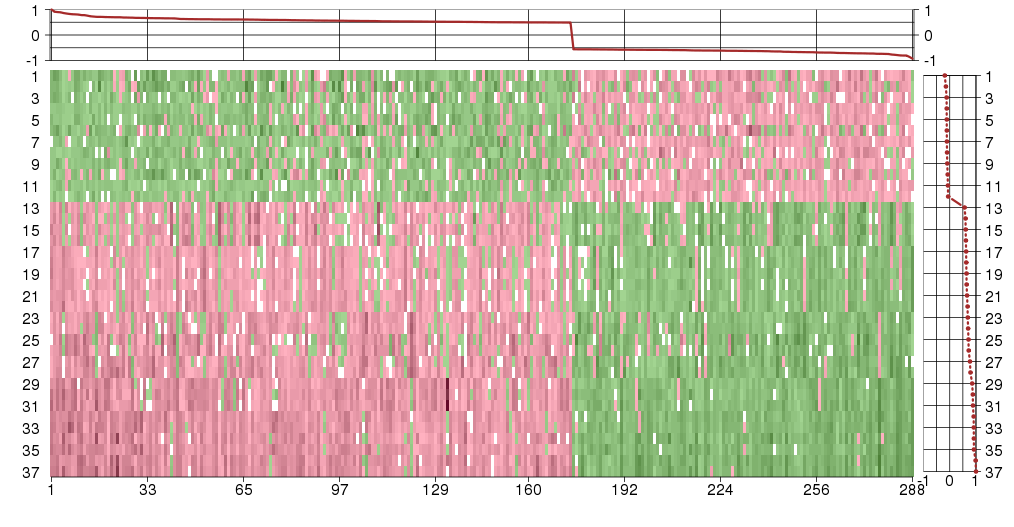

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
preribosome
Any complex of pre-rRNAs, ribosomal proteins, and associated proteins formed during ribosome biogenesis.
small-subunit processome
A large ribonucleoprotein complex that is an early preribosomal complex. In S. cerevisiae, it has a size of 80S and consists of the 35S pre-rRNA, early-associating ribosomal proteins most of which are part of the small ribosomal subunit, the U3 snoRNA and associated proteins.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.

AATFapoptosis antagonizing transcription factor (ENSG00000108270), score: -0.61 ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (ENSG00000165029), score: 0.49 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.7 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: 0.61 ABHD3abhydrolase domain containing 3 (ENSG00000158201), score: 0.61 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.76 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: 0.7 ADAP2ArfGAP with dual PH domains 2 (ENSG00000184060), score: 0.52 ADPRHADP-ribosylarginine hydrolase (ENSG00000144843), score: 0.53 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (ENSG00000117448), score: 0.56 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.57 ANGEL1angel homolog 1 (Drosophila) (ENSG00000013523), score: -0.59 ANGEL2angel homolog 2 (Drosophila) (ENSG00000174606), score: 0.56 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.67 ANKRD42ankyrin repeat domain 42 (ENSG00000137494), score: -0.56 AP2B1adaptor-related protein complex 2, beta 1 subunit (ENSG00000006125), score: -0.57 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.54 ARSBarylsulfatase B (ENSG00000113273), score: 0.59 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.49 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.61 ATL2atlastin GTPase 2 (ENSG00000119787), score: 0.53 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.67 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.77 ATP6V0A2ATPase, H+ transporting, lysosomal V0 subunit a2 (ENSG00000185344), score: 0.57 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: -0.63 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: 0.51 BEST1bestrophin 1 (ENSG00000167995), score: -0.6 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.67 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.67 C12orf5chromosome 12 open reading frame 5 (ENSG00000078237), score: 0.49 C13orf31chromosome 13 open reading frame 31 (ENSG00000179630), score: 0.54 C16orf80chromosome 16 open reading frame 80 (ENSG00000070761), score: -0.57 C17orf39chromosome 17 open reading frame 39 (ENSG00000141034), score: 0.62 C1orf103chromosome 1 open reading frame 103 (ENSG00000121931), score: -0.57 C1orf128chromosome 1 open reading frame 128 (ENSG00000057757), score: -0.61 C1orf27chromosome 1 open reading frame 27 (ENSG00000157181), score: 0.52 C1orf58chromosome 1 open reading frame 58 (ENSG00000162819), score: 0.49 C1orf83chromosome 1 open reading frame 83 (ENSG00000116205), score: 0.49 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.61 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.72 C21orf63chromosome 21 open reading frame 63 (ENSG00000166979), score: -0.57 C3orf59chromosome 3 open reading frame 59 (ENSG00000180611), score: 0.6 C9orf9chromosome 9 open reading frame 9 (ENSG00000165698), score: -0.61 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.86 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.71 CCDC99coiled-coil domain containing 99 (ENSG00000040275), score: -0.63 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.78 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.86 CENPNcentromere protein N (ENSG00000166451), score: -0.67 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.54 CHD1Lchromodomain helicase DNA binding protein 1-like (ENSG00000131778), score: -0.61 CHMP7CHMP family, member 7 (ENSG00000147457), score: 0.5 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.8 CLEC3BC-type lectin domain family 3, member B (ENSG00000163815), score: -0.58 CNOT6CCR4-NOT transcription complex, subunit 6 (ENSG00000113300), score: 0.59 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.7 CRKv-crk sarcoma virus CT10 oncogene homolog (avian) (ENSG00000167193), score: 0.53 CROTcarnitine O-octanoyltransferase (ENSG00000005469), score: 0.51 CTDP1CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 (ENSG00000060069), score: -0.63 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: 0.67 CYBRD1cytochrome b reductase 1 (ENSG00000071967), score: -0.58 DARSaspartyl-tRNA synthetase (ENSG00000115866), score: 0.6 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.61 DCTDdCMP deaminase (ENSG00000129187), score: -0.78 DDX41DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (ENSG00000183258), score: -0.7 DDX6DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (ENSG00000110367), score: 0.51 DERL1Der1-like domain family, member 1 (ENSG00000136986), score: 0.51 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: -0.73 DNALI1dynein, axonemal, light intermediate chain 1 (ENSG00000163879), score: -0.63 DPM1dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit (ENSG00000000419), score: 0.52 DTNBP1dystrobrevin binding protein 1 (ENSG00000047579), score: 0.53 EEA1early endosome antigen 1 (ENSG00000102189), score: 0.66 EFHC1EF-hand domain (C-terminal) containing 1 (ENSG00000096093), score: -0.58 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (ENSG00000172071), score: 0.61 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (ENSG00000158711), score: 0.52 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.81 ENTPD6ectonucleoside triphosphate diphosphohydrolase 6 (putative) (ENSG00000197586), score: -0.56 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: 0.52 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.67 ERN1endoplasmic reticulum to nucleus signaling 1 (ENSG00000178607), score: 0.49 ETFDHelectron-transferring-flavoprotein dehydrogenase (ENSG00000171503), score: 0.49 EVI5ecotropic viral integration site 5 (ENSG00000067208), score: 0.66 EXOC3exocyst complex component 3 (ENSG00000180104), score: -0.57 FAM105Bfamily with sequence similarity 105, member B (ENSG00000154124), score: 0.58 FAM118Bfamily with sequence similarity 118, member B (ENSG00000197798), score: -0.59 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: -0.64 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: 0.59 FAM199Xfamily with sequence similarity 199, X-linked (ENSG00000123575), score: -0.68 FAM20Bfamily with sequence similarity 20, member B (ENSG00000116199), score: 0.59 FAM36Afamily with sequence similarity 36, member A (ENSG00000203667), score: -0.58 FAM53Afamily with sequence similarity 53, member A (ENSG00000174137), score: 0.5 FAM76Afamily with sequence similarity 76, member A (ENSG00000009780), score: 0.53 FBXL7F-box and leucine-rich repeat protein 7 (ENSG00000183580), score: -0.59 FBXO22F-box protein 22 (ENSG00000167196), score: 0.6 FBXO3F-box protein 3 (ENSG00000110429), score: 0.78 FCHO2FCH domain only 2 (ENSG00000157107), score: 0.71 FIGNL1fidgetin-like 1 (ENSG00000132436), score: -0.61 FKTNfukutin (ENSG00000106692), score: 0.5 FRS2fibroblast growth factor receptor substrate 2 (ENSG00000166225), score: 0.51 GAPVD1GTPase activating protein and VPS9 domains 1 (ENSG00000165219), score: 0.57 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.61 GLIPR2GLI pathogenesis-related 2 (ENSG00000122694), score: -0.71 GPAMglycerol-3-phosphate acyltransferase, mitochondrial (ENSG00000119927), score: 0.49 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.62 GPX8glutathione peroxidase 8 (putative) (ENSG00000164294), score: 0.5 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: 0.72 H1FXH1 histone family, member X (ENSG00000184897), score: -0.74 HDDC2HD domain containing 2 (ENSG00000111906), score: -0.71 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.54 HMBOX1homeobox containing 1 (ENSG00000147421), score: 0.5 HNRNPRheterogeneous nuclear ribonucleoprotein R (ENSG00000125944), score: -0.64 HPRT1hypoxanthine phosphoribosyltransferase 1 (ENSG00000165704), score: 0.5 HSF1heat shock transcription factor 1 (ENSG00000185122), score: -0.72 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.61 ITM2Bintegral membrane protein 2B (ENSG00000136156), score: 0.58 JAK2Janus kinase 2 (ENSG00000096968), score: 0.66 KCNMB1potassium large conductance calcium-activated channel, subfamily M, beta member 1 (ENSG00000145936), score: -0.56 KCTD5potassium channel tetramerisation domain containing 5 (ENSG00000167977), score: -0.72 KIAA0146KIAA0146 (ENSG00000164808), score: -0.67 KIAA0174KIAA0174 (ENSG00000182149), score: -0.71 KIAA1033KIAA1033 (ENSG00000136051), score: 0.57 KIAA1370KIAA1370 (ENSG00000047346), score: 0.49 KIAA1432KIAA1432 (ENSG00000107036), score: 0.51 KIAA2018KIAA2018 (ENSG00000176542), score: 0.68 KLHL24kelch-like 24 (Drosophila) (ENSG00000114796), score: 0.49 LACE1lactation elevated 1 (ENSG00000135537), score: 0.65 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.7 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: 0.5 LIN7Clin-7 homolog C (C. elegans) (ENSG00000148943), score: 0.5 LMBRD2LMBR1 domain containing 2 (ENSG00000164187), score: 0.5 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: 0.58 LOC100302652GPR75-ASB3 (ENSG00000115239), score: -0.56 LPGAT1lysophosphatidylglycerol acyltransferase 1 (ENSG00000123684), score: 0.56 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 0.52 LSM10LSM10, U7 small nuclear RNA associated (ENSG00000181817), score: -0.61 MAD2L2MAD2 mitotic arrest deficient-like 2 (yeast) (ENSG00000116670), score: -0.74 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.62 MAP3K14mitogen-activated protein kinase kinase kinase 14 (ENSG00000006062), score: -0.63 MAPKSP1MAPK scaffold protein 1 (ENSG00000109270), score: 0.55 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.61 MASTLmicrotubule associated serine/threonine kinase-like (ENSG00000120539), score: -0.67 MBIPMAP3K12 binding inhibitory protein 1 (ENSG00000151332), score: -0.66 MCEEmethylmalonyl CoA epimerase (ENSG00000124370), score: 0.55 MCM8minichromosome maintenance complex component 8 (ENSG00000125885), score: -0.57 MDH1malate dehydrogenase 1, NAD (soluble) (ENSG00000014641), score: 0.52 MDM4Mdm4 p53 binding protein homolog (mouse) (ENSG00000198625), score: 0.59 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (ENSG00000082212), score: -0.65 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.58 MED1mediator complex subunit 1 (ENSG00000125686), score: 0.62 MFN1mitofusin 1 (ENSG00000171109), score: 0.81 MFSD10major facilitator superfamily domain containing 10 (ENSG00000109736), score: -0.58 MGST2microsomal glutathione S-transferase 2 (ENSG00000085871), score: -0.75 MIA3melanoma inhibitory activity family, member 3 (ENSG00000154305), score: 0.49 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.7 MMAAmethylmalonic aciduria (cobalamin deficiency) cblA type (ENSG00000151611), score: 0.55 MMEL1membrane metallo-endopeptidase-like 1 (ENSG00000142606), score: -0.69 MPP6membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (ENSG00000105926), score: 0.56 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: -0.58 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.55 NAA35N(alpha)-acetyltransferase 35, NatC auxiliary subunit (ENSG00000135040), score: 0.61 NAIF1nuclear apoptosis inducing factor 1 (ENSG00000171169), score: -0.71 NAPEPLDN-acyl phosphatidylethanolamine phospholipase D (ENSG00000161048), score: -0.59 ND3NADH dehydrogenase, subunit 3 (complex I) (ENSG00000198840), score: -0.6 NDUFA6NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa (ENSG00000184983), score: 0.65 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.8 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.64 NHEDC2Na+/H+ exchanger domain containing 2 (ENSG00000164038), score: -0.62 NINJ2ninjurin 2 (ENSG00000171840), score: -0.59 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.93 NSMAFneutral sphingomyelinase (N-SMase) activation associated factor (ENSG00000035681), score: 0.52 NT5C35'-nucleotidase, cytosolic III (ENSG00000122643), score: 0.62 NT5DC35'-nucleotidase domain containing 3 (ENSG00000111696), score: 0.57 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.83 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: 0.55 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.55 OSGIN2oxidative stress induced growth inhibitor family member 2 (ENSG00000164823), score: -0.69 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (ENSG00000122884), score: 0.5 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.61 PAK1IP1PAK1 interacting protein 1 (ENSG00000111845), score: 0.59 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: -0.69 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.55 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.55 PARP1poly (ADP-ribose) polymerase 1 (ENSG00000143799), score: -0.73 PASKPAS domain containing serine/threonine kinase (ENSG00000115687), score: -0.58 PCBD2pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 (ENSG00000132570), score: 0.6 PCID2PCI domain containing 2 (ENSG00000126226), score: -0.58 PDHBpyruvate dehydrogenase (lipoamide) beta (ENSG00000168291), score: 0.5 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.58 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.69 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.6 PIGPphosphatidylinositol glycan anchor biosynthesis, class P (ENSG00000185808), score: 0.61 PLEKHN1pleckstrin homology domain containing, family N member 1 (ENSG00000187583), score: 0.62 PMS1PMS1 postmeiotic segregation increased 1 (S. cerevisiae) (ENSG00000064933), score: -0.57 PNLDC1poly(A)-specific ribonuclease (PARN)-like domain containing 1 (ENSG00000146453), score: -0.58 POLLpolymerase (DNA directed), lambda (ENSG00000166169), score: -0.66 POLR2Cpolymerase (RNA) II (DNA directed) polypeptide C, 33kDa (ENSG00000102978), score: -0.59 PPARGC1Bperoxisome proliferator-activated receptor gamma, coactivator 1 beta (ENSG00000155846), score: 0.53 PPM1Bprotein phosphatase, Mg2+/Mn2+ dependent, 1B (ENSG00000138032), score: 0.62 PPP1R7protein phosphatase 1, regulatory (inhibitor) subunit 7 (ENSG00000115685), score: -0.69 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha (ENSG00000066027), score: 0.61 PPTC7PTC7 protein phosphatase homolog (S. cerevisiae) (ENSG00000196850), score: 0.51 PQLC1PQ loop repeat containing 1 (ENSG00000122490), score: -0.69 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.62 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (ENSG00000197170), score: 0.56 PSMG3proteasome (prosome, macropain) assembly chaperone 3 (ENSG00000157778), score: -0.74 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: 0.54 QSER1glutamine and serine rich 1 (ENSG00000060749), score: 0.53 RASD1RAS, dexamethasone-induced 1 (ENSG00000108551), score: -0.65 RBM14RNA binding motif protein 14 (ENSG00000239306), score: -0.66 RBM45RNA binding motif protein 45 (ENSG00000155636), score: 0.52 RC3H1ring finger and CCCH-type zinc finger domains 1 (ENSG00000135870), score: 0.5 RECQL4RecQ protein-like 4 (ENSG00000160957), score: -0.61 RFESDRieske (Fe-S) domain containing (ENSG00000175449), score: 0.49 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.65 RNF139ring finger protein 139 (ENSG00000170881), score: 0.61 RPRD2regulation of nuclear pre-mRNA domain containing 2 (ENSG00000163125), score: -0.61 RPS19BP1ribosomal protein S19 binding protein 1 (ENSG00000187051), score: -0.68 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.81 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.84 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.68 SBK1SH3-binding domain kinase 1 (ENSG00000188322), score: 0.57 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.62 SEP1515 kDa selenoprotein (ENSG00000183291), score: 0.49 SKP2S-phase kinase-associated protein 2 (p45) (ENSG00000145604), score: -0.58 SLC16A3solute carrier family 16, member 3 (monocarboxylic acid transporter 4) (ENSG00000141526), score: -0.62 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: 0.73 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (ENSG00000100372), score: 0.59 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: -0.59 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.58 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.53 SLC35B3solute carrier family 35, member B3 (ENSG00000124786), score: -0.56 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.71 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.79 SNRKSNF related kinase (ENSG00000163788), score: 0.62 SNX13sorting nexin 13 (ENSG00000071189), score: 0.66 SSBP1single-stranded DNA binding protein 1 (ENSG00000106028), score: -0.57 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.68 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: 0.52 STRN3striatin, calmodulin binding protein 3 (ENSG00000196792), score: 0.62 STYXL1serine/threonine/tyrosine interacting-like 1 (ENSG00000127952), score: -0.73 SUGP2SURP and G patch domain containing 2 (ENSG00000064607), score: -0.63 TADA3transcriptional adaptor 3 (ENSG00000171148), score: -0.64 TAF1ATATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa (ENSG00000143498), score: 0.67 TBC1D2TBC1 domain family, member 2 (ENSG00000095383), score: -0.58 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (ENSG00000133138), score: 0.51 TBCELtubulin folding cofactor E-like (ENSG00000154114), score: 0.63 TCFL5transcription factor-like 5 (basic helix-loop-helix) (ENSG00000101190), score: -0.81 TCOF1Treacher Collins-Franceschetti syndrome 1 (ENSG00000070814), score: -0.62 TCP11L2t-complex 11 (mouse)-like 2 (ENSG00000166046), score: 0.7 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (ENSG00000100726), score: -0.58 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.61 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.91 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.68 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.53 TMEM63Btransmembrane protein 63B (ENSG00000137216), score: 0.55 TMEM64transmembrane protein 64 (ENSG00000180694), score: 0.56 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.59 TOP3Atopoisomerase (DNA) III alpha (ENSG00000177302), score: -0.57 TRAF1TNF receptor-associated factor 1 (ENSG00000056558), score: -0.65 TTC33tetratricopeptide repeat domain 33 (ENSG00000113638), score: 0.49 TUBE1tubulin, epsilon 1 (ENSG00000074935), score: -0.74 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.67 TULP4tubby like protein 4 (ENSG00000130338), score: 0.49 TXNDC16thioredoxin domain containing 16 (ENSG00000087301), score: 0.53 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: 0.56 TXNRD1thioredoxin reductase 1 (ENSG00000198431), score: 0.5 TYW3tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) (ENSG00000162623), score: -0.58 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.9 UBXN11UBX domain protein 11 (ENSG00000158062), score: -0.7 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.89 UHRF1BP1LUHRF1 binding protein 1-like (ENSG00000111647), score: 0.5 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: 0.57 USP45ubiquitin specific peptidase 45 (ENSG00000123552), score: 0.52 UTP11LUTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) (ENSG00000183520), score: 0.56 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.63 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: 0.53 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: 0.6 WBP1WW domain binding protein 1 (ENSG00000115274), score: -0.72 WDR36WD repeat domain 36 (ENSG00000134987), score: 0.66 WDR89WD repeat domain 89 (ENSG00000140006), score: 1 WHSC2Wolf-Hirschhorn syndrome candidate 2 (ENSG00000185049), score: -0.59 WRBtryptophan rich basic protein (ENSG00000182093), score: -0.62 YPEL5yippee-like 5 (Drosophila) (ENSG00000119801), score: -0.56 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.65 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.71 ZCCHC8zinc finger, CCHC domain containing 8 (ENSG00000033030), score: 0.53 ZNF518Bzinc finger protein 518B (ENSG00000178163), score: -0.56 ZNHIT3zinc finger, HIT type 3 (ENSG00000108278), score: -0.62
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_ts_m1_ca1 | hsa | ts | m | 1 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_ts_m2_ca1 | hsa | ts | m | 2 |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| ppa_ts_m_ca1 | ppa | ts | m | _ |
| ptr_br_m2_ca1 | ptr | br | m | 2 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |
| ptr_br_m3_ca1 | ptr | br | m | 3 |
| ggo_br_m_ca1 | ggo | br | m | _ |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_br_f_ca1 | mdo | br | f | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mmu_br_m1_ca1 | mmu | br | m | 1 |
| mmu_br_m2_ca1 | mmu | br | m | 2 |
| mmu_cb_m1_ca1 | mmu | cb | m | 1 |
| mmu_br_f_ca1 | mmu | br | f | _ |
| mmu_cb_f_ca1 | mmu | cb | f | _ |
| mmu_cb_m2_ca1 | mmu | cb | m | 2 |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |