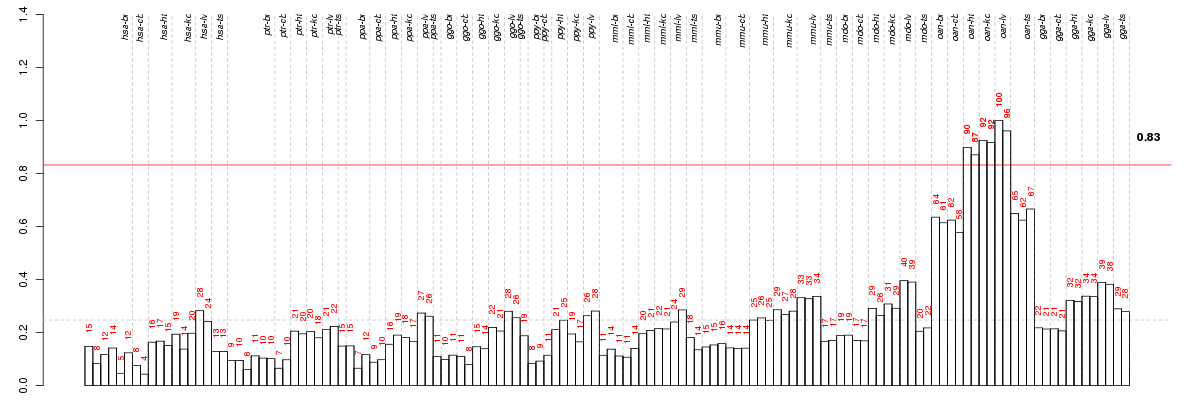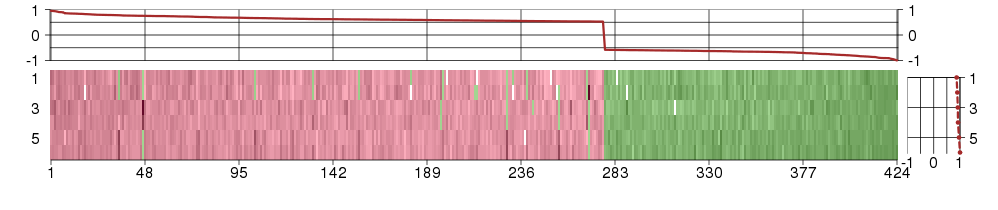

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
lytic vacuole
A vacuole that is maintained at an acidic pH and which contains degradative enzymes, including a wide variety of acid hydrolases.
lysosome
A small lytic vacuole that has cell cycle-independent morphology and is found in most animal cells and that contains a variety of hydrolases, most of which have their maximal activities in the pH range 5-6. The contained enzymes display latency if properly isolated. About 40 different lysosomal hydrolases are known and lysosomes have a great variety of morphologies and functions.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.

AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.76 AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: 0.59 ABCB7ATP-binding cassette, sub-family B (MDR/TAP), member 7 (ENSG00000131269), score: 0.6 ABHD12abhydrolase domain containing 12 (ENSG00000100997), score: -0.61 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.84 ACTN4actinin, alpha 4 (ENSG00000130402), score: -0.61 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: -0.6 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.71 ADAadenosine deaminase (ENSG00000196839), score: 0.63 ADALadenosine deaminase-like (ENSG00000168803), score: 0.66 ADSLadenylosuccinate lyase (ENSG00000239900), score: 0.62 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.54 AMOTL1angiomotin like 1 (ENSG00000166025), score: -0.59 AMPD1adenosine monophosphate deaminase 1 (ENSG00000116748), score: 0.53 ANAPC2anaphase promoting complex subunit 2 (ENSG00000176248), score: -0.68 ANKLE2ankyrin repeat and LEM domain containing 2 (ENSG00000176915), score: -0.65 ANKRD27ankyrin repeat domain 27 (VPS9 domain) (ENSG00000105186), score: -0.58 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: -0.8 AP3M2adaptor-related protein complex 3, mu 2 subunit (ENSG00000070718), score: -0.68 AQRaquarius homolog (mouse) (ENSG00000021776), score: 0.8 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (ENSG00000101199), score: -0.84 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: 0.58 ARHGEF18Rho/Rac guanine nucleotide exchange factor (GEF) 18 (ENSG00000104880), score: -0.62 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (ENSG00000163947), score: -0.66 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.58 ARL5BADP-ribosylation factor-like 5B (ENSG00000165997), score: 0.68 ARMC8armadillo repeat containing 8 (ENSG00000114098), score: 0.58 ASAH1N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ENSG00000104763), score: 0.61 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ENSG00000153317), score: -0.59 ASB13ankyrin repeat and SOCS box-containing 13 (ENSG00000196372), score: -0.63 ASF1AASF1 anti-silencing function 1 homolog A (S. cerevisiae) (ENSG00000111875), score: 0.74 ASH2Lash2 (absent, small, or homeotic)-like (Drosophila) (ENSG00000129691), score: -0.75 ATF6activating transcription factor 6 (ENSG00000118217), score: 0.62 ATG13ATG13 autophagy related 13 homolog (S. cerevisiae) (ENSG00000175224), score: 0.7 ATP10AATPase, class V, type 10A (ENSG00000206190), score: -0.59 ATPAF2ATP synthase mitochondrial F1 complex assembly factor 2 (ENSG00000171953), score: 0.61 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.55 AZI15-azacytidine induced 1 (ENSG00000141577), score: -0.74 B3GALNT1beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) (ENSG00000169255), score: -0.62 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (ENSG00000158470), score: -0.85 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (ENSG00000083123), score: 0.66 BCL2L13BCL2-like 13 (apoptosis facilitator) (ENSG00000099968), score: 0.62 BDKRB1bradykinin receptor B1 (ENSG00000100739), score: 0.63 BICD2bicaudal D homolog 2 (Drosophila) (ENSG00000185963), score: -0.6 BTKBruton agammaglobulinemia tyrosine kinase (ENSG00000010671), score: 0.58 BTRCbeta-transducin repeat containing (ENSG00000166167), score: -0.63 C12orf26chromosome 12 open reading frame 26 (ENSG00000127720), score: 0.76 C12orf65chromosome 12 open reading frame 65 (ENSG00000130921), score: -0.58 C14orf101chromosome 14 open reading frame 101 (ENSG00000070269), score: 0.69 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.78 C14orf126chromosome 14 open reading frame 126 (ENSG00000129480), score: 0.61 C16orf7chromosome 16 open reading frame 7 (ENSG00000075399), score: -0.65 C17orf103chromosome 17 open reading frame 103 (ENSG00000154035), score: 0.62 C17orf56chromosome 17 open reading frame 56 (ENSG00000167302), score: -0.7 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: -0.67 C17orf71chromosome 17 open reading frame 71 (ENSG00000167447), score: 0.53 C18orf8chromosome 18 open reading frame 8 (ENSG00000141452), score: 0.74 C19orf44chromosome 19 open reading frame 44 (ENSG00000105072), score: -0.59 C1orf25chromosome 1 open reading frame 25 (ENSG00000121486), score: 0.68 C1orf83chromosome 1 open reading frame 83 (ENSG00000116205), score: 0.65 C20orf4chromosome 20 open reading frame 4 (ENSG00000131043), score: 0.79 C5orf43chromosome 5 open reading frame 43 (ENSG00000188725), score: 0.6 C6orf57chromosome 6 open reading frame 57 (ENSG00000154079), score: 0.55 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: 0.9 C6orf89chromosome 6 open reading frame 89 (ENSG00000198663), score: 0.66 C8orf40chromosome 8 open reading frame 40 (ENSG00000176209), score: -0.84 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.54 CAND2cullin-associated and neddylation-dissociated 2 (putative) (ENSG00000144712), score: -0.58 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: 0.6 CCDC127coiled-coil domain containing 127 (ENSG00000164366), score: -0.83 CCDC28Acoiled-coil domain containing 28A (ENSG00000024862), score: -0.72 CCDC93coiled-coil domain containing 93 (ENSG00000125633), score: 0.63 CCNHcyclin H (ENSG00000134480), score: -0.59 CCNIcyclin I (ENSG00000118816), score: 0.61 CD86CD86 molecule (ENSG00000114013), score: 0.74 CDC45cell division cycle 45 homolog (S. cerevisiae) (ENSG00000093009), score: 0.68 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.93 CDT1chromatin licensing and DNA replication factor 1 (ENSG00000167513), score: 0.53 CENPQcentromere protein Q (ENSG00000031691), score: 0.57 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.55 CHMP6chromatin modifying protein 6 (ENSG00000176108), score: 0.7 CHRNA9cholinergic receptor, nicotinic, alpha 9 (ENSG00000174343), score: 0.53 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.74 CLK4CDC-like kinase 4 (ENSG00000113240), score: -0.82 CLPXClpX caseinolytic peptidase X homolog (E. coli) (ENSG00000166855), score: 0.78 CLTAclathrin, light chain A (ENSG00000122705), score: -0.65 CNOT2CCR4-NOT transcription complex, subunit 2 (ENSG00000111596), score: -0.62 COG2component of oligomeric golgi complex 2 (ENSG00000135775), score: 0.54 COG4component of oligomeric golgi complex 4 (ENSG00000103051), score: -0.63 COG7component of oligomeric golgi complex 7 (ENSG00000168434), score: 0.56 COMMD2COMM domain containing 2 (ENSG00000114744), score: 0.6 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -0.97 COPS7ACOP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) (ENSG00000111652), score: 0.67 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (ENSG00000173085), score: 0.71 COQ3coenzyme Q3 homolog, methyltransferase (S. cerevisiae) (ENSG00000132423), score: 0.56 COX18COX18 cytochrome c oxidase assembly homolog (S. cerevisiae) (ENSG00000163626), score: 0.54 CRLS1cardiolipin synthase 1 (ENSG00000088766), score: 0.57 CSE1LCSE1 chromosome segregation 1-like (yeast) (ENSG00000124207), score: -0.73 CTCFCCCTC-binding factor (zinc finger protein) (ENSG00000102974), score: -0.61 CUEDC2CUE domain containing 2 (ENSG00000107874), score: -0.66 CWF19L1CWF19-like 1, cell cycle control (S. pombe) (ENSG00000095485), score: 0.56 CXorf23chromosome X open reading frame 23 (ENSG00000173681), score: 0.58 CXXC5CXXC finger 5 (ENSG00000171604), score: -0.63 CYSLTR1cysteinyl leukotriene receptor 1 (ENSG00000173198), score: 0.85 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.6 DCTN4dynactin 4 (p62) (ENSG00000132912), score: 0.54 DDA1DET1 and DDB1 associated 1 (ENSG00000130311), score: -0.61 DENND1BDENN/MADD domain containing 1B (ENSG00000213047), score: 0.54 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.54 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: 0.62 DIRC2disrupted in renal carcinoma 2 (ENSG00000138463), score: 0.89 DLDdihydrolipoamide dehydrogenase (ENSG00000091140), score: 0.55 DNTTIP1deoxynucleotidyltransferase, terminal, interacting protein 1 (ENSG00000101457), score: 0.9 DONSONdownstream neighbor of SON (ENSG00000159147), score: 0.59 DPP8dipeptidyl-peptidase 8 (ENSG00000074603), score: 0.74 DRAM2DNA-damage regulated autophagy modulator 2 (ENSG00000156171), score: 0.59 DSCC1defective in sister chromatid cohesion 1 homolog (S. cerevisiae) (ENSG00000136982), score: 0.76 DSCR3Down syndrome critical region gene 3 (ENSG00000157538), score: 0.68 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.7 EDC3enhancer of mRNA decapping 3 homolog (S. cerevisiae) (ENSG00000179151), score: 0.65 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.57 EEF1E1eukaryotic translation elongation factor 1 epsilon 1 (ENSG00000124802), score: -0.65 EIF2B4eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa (ENSG00000115211), score: -0.59 EIF3Deukaryotic translation initiation factor 3, subunit D (ENSG00000100353), score: 0.53 EIF4E3eukaryotic translation initiation factor 4E family member 3 (ENSG00000163412), score: -0.6 ERLEC1endoplasmic reticulum lectin 1 (ENSG00000068912), score: 0.67 EXO1exonuclease 1 (ENSG00000174371), score: 0.64 EXOC8exocyst complex component 8 (ENSG00000116903), score: 0.73 EXOSC1exosome component 1 (ENSG00000171311), score: 0.59 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.65 FAM185Afamily with sequence similarity 185, member A (ENSG00000222011), score: -0.73 FAM63Bfamily with sequence similarity 63, member B (ENSG00000128923), score: 0.84 FAM82Bfamily with sequence similarity 82, member B (ENSG00000176623), score: 0.53 FAM96Afamily with sequence similarity 96, member A (ENSG00000166797), score: 0.56 FARP2FERM, RhoGEF and pleckstrin domain protein 2 (ENSG00000006607), score: -0.61 FBXO21F-box protein 21 (ENSG00000135108), score: -0.68 FBXO4F-box protein 4 (ENSG00000151876), score: 0.69 FHfumarate hydratase (ENSG00000091483), score: 0.71 FIGNL1fidgetin-like 1 (ENSG00000132436), score: 0.58 FSHBfollicle stimulating hormone, beta polypeptide (ENSG00000131808), score: 0.75 FTSJD2FtsJ methyltransferase domain containing 2 (ENSG00000137200), score: -0.7 FZD2frizzled homolog 2 (Drosophila) (ENSG00000180340), score: 0.56 GABPAGA binding protein transcription factor, alpha subunit 60kDa (ENSG00000154727), score: 0.57 GABPB1GA binding protein transcription factor, beta subunit 1 (ENSG00000104064), score: 0.57 GALK2galactokinase 2 (ENSG00000156958), score: 0.54 GAR1GAR1 ribonucleoprotein homolog (yeast) (ENSG00000109534), score: 0.57 GAS8growth arrest-specific 8 (ENSG00000141013), score: -0.65 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.61 GGHgamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (ENSG00000137563), score: 0.53 GIN1gypsy retrotransposon integrase 1 (ENSG00000145723), score: -0.61 GMEB2glucocorticoid modulatory element binding protein 2 (ENSG00000101216), score: -0.62 GOLGA1golgin A1 (ENSG00000136935), score: -0.78 GOLIM4golgi integral membrane protein 4 (ENSG00000173905), score: 0.54 GOLT1Bgolgi transport 1B (ENSG00000111711), score: 0.6 GORASP1golgi reassembly stacking protein 1, 65kDa (ENSG00000114745), score: 0.6 GOT1glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (ENSG00000120053), score: 0.53 GPN1GPN-loop GTPase 1 (ENSG00000198522), score: 0.94 GTF2E1general transcription factor IIE, polypeptide 1, alpha 56kDa (ENSG00000153767), score: 0.76 GTPBP8GTP-binding protein 8 (putative) (ENSG00000163607), score: 0.6 HEBP1heme binding protein 1 (ENSG00000013583), score: 0.63 HELQhelicase, POLQ-like (ENSG00000163312), score: 0.73 HERPUD2HERPUD family member 2 (ENSG00000122557), score: 0.58 HM13histocompatibility (minor) 13 (ENSG00000101294), score: 0.55 HMGXB4HMG box domain containing 4 (ENSG00000100281), score: 0.72 HSPB11heat shock protein family B (small), member 11 (ENSG00000081870), score: 0.76 HTThuntingtin (ENSG00000197386), score: -0.91 HYIhydroxypyruvate isomerase (putative) (ENSG00000178922), score: 0.59 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (ENSG00000005700), score: 0.68 IDEinsulin-degrading enzyme (ENSG00000119912), score: 0.65 INTS9integrator complex subunit 9 (ENSG00000104299), score: 0.64 IPO11importin 11 (ENSG00000086200), score: 0.69 IQCB1IQ motif containing B1 (ENSG00000173226), score: -0.81 ITCHitchy E3 ubiquitin protein ligase homolog (mouse) (ENSG00000078747), score: 0.53 ITGA4integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) (ENSG00000115232), score: 0.64 ITGA8integrin, alpha 8 (ENSG00000077943), score: 0.6 ITPK1inositol 1,3,4-triphosphate 5/6 kinase (ENSG00000100605), score: -0.59 JOSD1Josephin domain containing 1 (ENSG00000100221), score: -0.59 KBTBD2kelch repeat and BTB (POZ) domain containing 2 (ENSG00000170852), score: 0.53 KDELC1KDEL (Lys-Asp-Glu-Leu) containing 1 (ENSG00000134901), score: 0.56 KDM1Blysine (K)-specific demethylase 1B (ENSG00000165097), score: 0.61 KIAA0090KIAA0090 (ENSG00000127463), score: 0.72 KIAA0226KIAA0226 (ENSG00000145016), score: -0.67 KIAA0247KIAA0247 (ENSG00000100647), score: 0.58 KIAA0664KIAA0664 (ENSG00000132361), score: 0.55 KIAA0907KIAA0907 (ENSG00000132680), score: 0.95 KIAA1407KIAA1407 (ENSG00000163617), score: 0.73 KLHDC8Bkelch domain containing 8B (ENSG00000185909), score: -0.63 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.63 KLHL7kelch-like 7 (Drosophila) (ENSG00000122550), score: -0.66 KLHL8kelch-like 8 (Drosophila) (ENSG00000145332), score: 0.76 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (ENSG00000117009), score: 0.61 KRIT1KRIT1, ankyrin repeat containing (ENSG00000001631), score: 0.58 LACTBlactamase, beta (ENSG00000103642), score: 0.61 LAMB3laminin, beta 3 (ENSG00000196878), score: 0.62 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.65 LEAP2liver expressed antimicrobial peptide 2 (ENSG00000164406), score: 0.63 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: -0.63 LOC100134387similar to DEAH (Asp-Glu-Ala-His) box polypeptide 40 (ENSG00000108406), score: -0.65 LOC100292021similar to thioredoxin peroxidase (ENSG00000123131), score: 0.58 LOC100302652GPR75-ASB3 (ENSG00000115239), score: 0.84 LOC653155PRP4 pre-mRNA processing factor 4 homolog B (yeast) pseudogene (ENSG00000112739), score: -0.63 LOH12CR1loss of heterozygosity, 12, chromosomal region 1 (ENSG00000165714), score: 0.74 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (ENSG00000126243), score: -0.62 LRIG3leucine-rich repeats and immunoglobulin-like domains 3 (ENSG00000139263), score: 0.68 LRP11low density lipoprotein receptor-related protein 11 (ENSG00000120256), score: -0.67 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.56 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.66 LRRC58leucine rich repeat containing 58 (ENSG00000163428), score: 0.69 MAGT1magnesium transporter 1 (ENSG00000102158), score: 0.53 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: 0.81 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: 0.57 MAP2K5mitogen-activated protein kinase kinase 5 (ENSG00000137764), score: -0.79 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (ENSG00000138834), score: -0.68 MBD3methyl-CpG binding domain protein 3 (ENSG00000071655), score: -0.59 MBNL2muscleblind-like 2 (Drosophila) (ENSG00000139793), score: -0.95 MCM6minichromosome maintenance complex component 6 (ENSG00000076003), score: 0.64 MED23mediator complex subunit 23 (ENSG00000112282), score: 0.78 MLLT1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 (ENSG00000130382), score: -0.66 MLXMAX-like protein X (ENSG00000108788), score: 0.57 MOCS2molybdenum cofactor synthesis 2 (ENSG00000164172), score: 0.58 MPDZmultiple PDZ domain protein (ENSG00000107186), score: -0.7 MRPL12mitochondrial ribosomal protein L12 (ENSG00000183093), score: -0.84 MRPL17mitochondrial ribosomal protein L17 (ENSG00000158042), score: 0.55 MRPL44mitochondrial ribosomal protein L44 (ENSG00000135900), score: 0.6 MRPS11mitochondrial ribosomal protein S11 (ENSG00000181991), score: 0.6 MRPS18Amitochondrial ribosomal protein S18A (ENSG00000096080), score: 0.72 MRPS30mitochondrial ribosomal protein S30 (ENSG00000112996), score: 0.6 MRPS35mitochondrial ribosomal protein S35 (ENSG00000061794), score: 0.56 MSRAmethionine sulfoxide reductase A (ENSG00000175806), score: 0.64 MTA1metastasis associated 1 (ENSG00000182979), score: -0.86 MTAPmethylthioadenosine phosphorylase (ENSG00000099810), score: 0.83 MTPAPmitochondrial poly(A) polymerase (ENSG00000107951), score: 0.62 MYBBP1AMYB binding protein (P160) 1a (ENSG00000132382), score: 0.55 MYLIPmyosin regulatory light chain interacting protein (ENSG00000007944), score: 0.59 MYNNmyoneurin (ENSG00000085274), score: 0.85 N6AMT2N-6 adenine-specific DNA methyltransferase 2 (putative) (ENSG00000150456), score: 0.67 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.58 NARS2asparaginyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000137513), score: 0.54 NCSTNnicastrin (ENSG00000162736), score: 0.63 NEDD1neural precursor cell expressed, developmentally down-regulated 1 (ENSG00000139350), score: 0.53 NELFnasal embryonic LHRH factor (ENSG00000165802), score: -0.64 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: 0.71 NPAS2neuronal PAS domain protein 2 (ENSG00000170485), score: -0.6 NPATnuclear protein, ataxia-telangiectasia locus (ENSG00000149308), score: 0.61 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.75 NRBP1nuclear receptor binding protein 1 (ENSG00000115216), score: -0.77 NRP2neuropilin 2 (ENSG00000118257), score: 0.54 NT5DC25'-nucleotidase domain containing 2 (ENSG00000168268), score: -0.74 NT5E5'-nucleotidase, ecto (CD73) (ENSG00000135318), score: 0.75 NUP133nucleoporin 133kDa (ENSG00000069248), score: 0.62 NUP37nucleoporin 37kDa (ENSG00000075188), score: 0.71 OLFML2Bolfactomedin-like 2B (ENSG00000162745), score: 0.61 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: -0.62 OSTM1osteopetrosis associated transmembrane protein 1 (ENSG00000081087), score: 0.69 OTCornithine carbamoyltransferase (ENSG00000036473), score: 0.55 OXSR1oxidative-stress responsive 1 (ENSG00000172939), score: 0.78 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: -0.68 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.6 PAIP2poly(A) binding protein interacting protein 2 (ENSG00000120727), score: 0.55 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: 0.53 PARP6poly (ADP-ribose) polymerase family, member 6 (ENSG00000137817), score: -0.88 PCGF3polycomb group ring finger 3 (ENSG00000185619), score: -0.6 PDE3Bphosphodiesterase 3B, cGMP-inhibited (ENSG00000152270), score: 0.53 PDRG1p53 and DNA-damage regulated 1 (ENSG00000088356), score: 0.77 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.65 PGM3phosphoglucomutase 3 (ENSG00000013375), score: 0.93 PHF16PHD finger protein 16 (ENSG00000102221), score: 0.59 PHOSPHO1phosphatase, orphan 1 (ENSG00000173868), score: 0.67 PIAS1protein inhibitor of activated STAT, 1 (ENSG00000033800), score: -0.59 PIGXphosphatidylinositol glycan anchor biosynthesis, class X (ENSG00000163964), score: 0.8 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (ENSG00000186111), score: -0.72 PITPNAphosphatidylinositol transfer protein, alpha (ENSG00000174238), score: -0.62 PLDNpallidin homolog (mouse) (ENSG00000104164), score: 0.73 PLEKHF2pleckstrin homology domain containing, family F (with FYVE domain) member 2 (ENSG00000175895), score: 0.62 PM20D2peptidase M20 domain containing 2 (ENSG00000146281), score: 0.69 PNPLA6patatin-like phospholipase domain containing 6 (ENSG00000032444), score: -0.6 POFUT1protein O-fucosyltransferase 1 (ENSG00000101346), score: 0.62 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (ENSG00000132664), score: 0.83 PPIL3peptidylprolyl isomerase (cyclophilin)-like 3 (ENSG00000240344), score: 0.67 PPP1R8protein phosphatase 1, regulatory (inhibitor) subunit 8 (ENSG00000117751), score: -0.66 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta (ENSG00000175470), score: -0.65 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta (ENSG00000112640), score: -0.74 PRKCDprotein kinase C, delta (ENSG00000163932), score: -0.68 PRMT10protein arginine methyltransferase 10 (putative) (ENSG00000164169), score: 0.56 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (ENSG00000101161), score: -0.67 PSMA3proteasome (prosome, macropain) subunit, alpha type, 3 (ENSG00000100567), score: 0.61 PSMG3proteasome (prosome, macropain) assembly chaperone 3 (ENSG00000157778), score: 0.53 PTPN9protein tyrosine phosphatase, non-receptor type 9 (ENSG00000169410), score: -0.61 QPCTglutaminyl-peptide cyclotransferase (ENSG00000115828), score: 0.58 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.85 RAB33BRAB33B, member RAS oncogene family (ENSG00000172007), score: 0.64 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.57 RAD17RAD17 homolog (S. pombe) (ENSG00000152942), score: 0.62 RAD54L2RAD54-like 2 (S. cerevisiae) (ENSG00000164080), score: -0.62 RAI1retinoic acid induced 1 (ENSG00000108557), score: -0.6 RANBP3RAN binding protein 3 (ENSG00000031823), score: -0.86 RBM45RNA binding motif protein 45 (ENSG00000155636), score: 0.56 RCHY1ring finger and CHY zinc finger domain containing 1 (ENSG00000163743), score: 0.63 RCOR3REST corepressor 3 (ENSG00000117625), score: -0.61 RECKreversion-inducing-cysteine-rich protein with kazal motifs (ENSG00000122707), score: 0.81 RFC3replication factor C (activator 1) 3, 38kDa (ENSG00000133119), score: 0.59 RG9MTD1RNA (guanine-9-) methyltransferase domain containing 1 (ENSG00000174173), score: 0.71 RNF103ring finger protein 103 (ENSG00000239305), score: 0.54 RNF144Aring finger protein 144A (ENSG00000151692), score: -0.64 RNF185ring finger protein 185 (ENSG00000138942), score: -0.78 RNF34ring finger protein 34 (ENSG00000170633), score: -0.61 RNF38ring finger protein 38 (ENSG00000137075), score: -0.61 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: 0.61 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: -0.66 ROBLD3roadblock domain containing 3 (ENSG00000116586), score: 0.59 RORARAR-related orphan receptor A (ENSG00000069667), score: -0.66 RPAINRPA interacting protein (ENSG00000129197), score: 0.84 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: 0.59 SAMD8sterile alpha motif domain containing 8 (ENSG00000156671), score: -0.61 SCLYselenocysteine lyase (ENSG00000132330), score: -0.59 SDCCAG8serologically defined colon cancer antigen 8 (ENSG00000054282), score: -0.64 SERINC5serine incorporator 5 (ENSG00000164300), score: 0.66 SERPINB5serpin peptidase inhibitor, clade B (ovalbumin), member 5 (ENSG00000206075), score: 0.53 SETD5SET domain containing 5 (ENSG00000168137), score: -1 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: -0.62 SIRT3sirtuin 3 (ENSG00000142082), score: 0.6 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (ENSG00000204351), score: 0.72 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (ENSG00000119899), score: 0.59 SLC30A1solute carrier family 30 (zinc transporter), member 1 (ENSG00000170385), score: 0.53 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.54 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.56 SLC39A9solute carrier family 39 (zinc transporter), member 9 (ENSG00000029364), score: 0.82 SNAPC1small nuclear RNA activating complex, polypeptide 1, 43kDa (ENSG00000023608), score: 0.56 SNED1sushi, nidogen and EGF-like domains 1 (ENSG00000162804), score: -0.81 SNRNP48small nuclear ribonucleoprotein 48kDa (U11/U12) (ENSG00000168566), score: 0.75 SNX24sorting nexin 24 (ENSG00000064652), score: 0.66 SPATA5L1spermatogenesis associated 5-like 1 (ENSG00000171763), score: 0.75 SPENspen homolog, transcriptional regulator (Drosophila) (ENSG00000065526), score: -0.86 SPOPLspeckle-type POZ protein-like (ENSG00000144228), score: 0.55 SPPL2Bsignal peptide peptidase-like 2B (ENSG00000005206), score: -0.67 SRSF4serine/arginine-rich splicing factor 4 (ENSG00000116350), score: -0.73 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: -0.9 SS18synovial sarcoma translocation, chromosome 18 (ENSG00000141380), score: 0.61 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: 0.56 ST6GALNAC6ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 (ENSG00000160408), score: -0.58 ST8SIA4ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (ENSG00000113532), score: 0.67 STK24serine/threonine kinase 24 (ENSG00000102572), score: -0.66 STK39serine threonine kinase 39 (ENSG00000198648), score: -0.63 STOML1stomatin (EPB72)-like 1 (ENSG00000067221), score: -0.6 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.91 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: 0.75 SURF1surfeit 1 (ENSG00000148290), score: 0.66 TAB3TGF-beta activated kinase 1/MAP3K7 binding protein 3 (ENSG00000157625), score: 0.65 TARSL2threonyl-tRNA synthetase-like 2 (ENSG00000185418), score: -0.84 TBC1D2BTBC1 domain family, member 2B (ENSG00000167202), score: -0.61 TBCCtubulin folding cofactor C (ENSG00000124659), score: 0.61 TBCEtubulin folding cofactor E (ENSG00000116957), score: 0.74 TCEA2transcription elongation factor A (SII), 2 (ENSG00000171703), score: -0.65 TCF12transcription factor 12 (ENSG00000140262), score: 0.79 TDRD7tudor domain containing 7 (ENSG00000196116), score: -0.79 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.63 THUMPD2THUMP domain containing 2 (ENSG00000138050), score: 0.57 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: 0.57 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.72 TMCC1transmembrane and coiled-coil domain family 1 (ENSG00000172765), score: -0.78 TMCO3transmembrane and coiled-coil domains 3 (ENSG00000150403), score: -0.68 TMEM110transmembrane protein 110 (ENSG00000213533), score: -0.65 TMEM18transmembrane protein 18 (ENSG00000151353), score: 0.53 TMEM33transmembrane protein 33 (ENSG00000109133), score: 0.76 TMEM60transmembrane protein 60 (ENSG00000135211), score: 0.75 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.76 TMEM9BTMEM9 domain family, member B (ENSG00000175348), score: 0.55 TMX4thioredoxin-related transmembrane protein 4 (ENSG00000125827), score: -0.6 TOM1L1target of myb1 (chicken)-like 1 (ENSG00000141198), score: -0.6 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: -0.63 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.58 TPP2tripeptidyl peptidase II (ENSG00000134900), score: 0.54 TPST1tyrosylprotein sulfotransferase 1 (ENSG00000169902), score: 0.62 TRAF3IP1TNF receptor-associated factor 3 interacting protein 1 (ENSG00000204104), score: -0.66 TRAPPC3trafficking protein particle complex 3 (ENSG00000054116), score: -0.64 TRIM23tripartite motif-containing 23 (ENSG00000113595), score: 0.58 TRIM7tripartite motif-containing 7 (ENSG00000146054), score: 0.61 TRIP13thyroid hormone receptor interactor 13 (ENSG00000071539), score: 0.79 TRNAU1APtRNA selenocysteine 1 associated protein 1 (ENSG00000180098), score: 0.68 TSG101tumor susceptibility gene 101 (ENSG00000074319), score: 0.63 TSNtranslin (ENSG00000211460), score: 0.66 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: -0.76 TTC1tetratricopeptide repeat domain 1 (ENSG00000113312), score: -0.72 TTC13tetratricopeptide repeat domain 13 (ENSG00000143643), score: 0.75 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.89 TWISTNBTWIST neighbor (ENSG00000105849), score: 0.64 TXNDC15thioredoxin domain containing 15 (ENSG00000113621), score: 0.74 UBA6ubiquitin-like modifier activating enzyme 6 (ENSG00000033178), score: 0.68 UBE2CBPubiquitin-conjugating enzyme E2C binding protein (ENSG00000118420), score: 0.73 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: 0.62 UBL3ubiquitin-like 3 (ENSG00000122042), score: 0.79 UBXN4UBX domain protein 4 (ENSG00000144224), score: 0.69 UIMC1ubiquitin interaction motif containing 1 (ENSG00000087206), score: -0.63 UNC50unc-50 homolog (C. elegans) (ENSG00000115446), score: 0.66 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: 0.54 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: -0.75 USP20ubiquitin specific peptidase 20 (ENSG00000136878), score: -0.8 UTP15UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) (ENSG00000164338), score: 0.79 UTP23UTP23, small subunit (SSU) processome component, homolog (yeast) (ENSG00000147679), score: 0.6 VAT1vesicle amine transport protein 1 homolog (T. californica) (ENSG00000108828), score: 0.73 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.92 VPS4Bvacuolar protein sorting 4 homolog B (S. cerevisiae) (ENSG00000119541), score: 0.78 VTA1Vps20-associated 1 homolog (S. cerevisiae) (ENSG00000009844), score: 0.56 WASF3WAS protein family, member 3 (ENSG00000132970), score: -0.6 WDR3WD repeat domain 3 (ENSG00000065183), score: 0.82 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.71 WDR6WD repeat domain 6 (ENSG00000178252), score: -0.58 WDR76WD repeat domain 76 (ENSG00000092470), score: 0.74 WDR8WD repeat domain 8 (ENSG00000116213), score: -0.71 WNT9Awingless-type MMTV integration site family, member 9A (ENSG00000143816), score: 0.57 XKR9XK, Kell blood group complex subunit-related family, member 9 (ENSG00000221947), score: 0.61 XRCC6BP1XRCC6 binding protein 1 (ENSG00000166896), score: 0.83 YIPF1Yip1 domain family, member 1 (ENSG00000058799), score: 0.65 YTHDC2YTH domain containing 2 (ENSG00000047188), score: -0.9 ZBTB1zinc finger and BTB domain containing 1 (ENSG00000126804), score: 0.56 ZBTB24zinc finger and BTB domain containing 24 (ENSG00000112365), score: 0.63 ZBTB49zinc finger and BTB domain containing 49 (ENSG00000168826), score: -0.9 ZBTB8Azinc finger and BTB domain containing 8A (ENSG00000160062), score: 0.59 ZC3H12Dzinc finger CCCH-type containing 12D (ENSG00000178199), score: 0.78 ZHX1zinc fingers and homeoboxes 1 (ENSG00000165156), score: 0.54 ZMPSTE24zinc metallopeptidase (STE24 homolog, S. cerevisiae) (ENSG00000084073), score: 0.75 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.78 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.75 ZW10ZW10, kinetochore associated, homolog (Drosophila) (ENSG00000086827), score: 0.79
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_ht_f_ca1 | oan | ht | f | _ |
| oan_ht_m_ca1 | oan | ht | m | _ |
| oan_kd_f_ca1 | oan | kd | f | _ |
| oan_kd_m_ca1 | oan | kd | m | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |