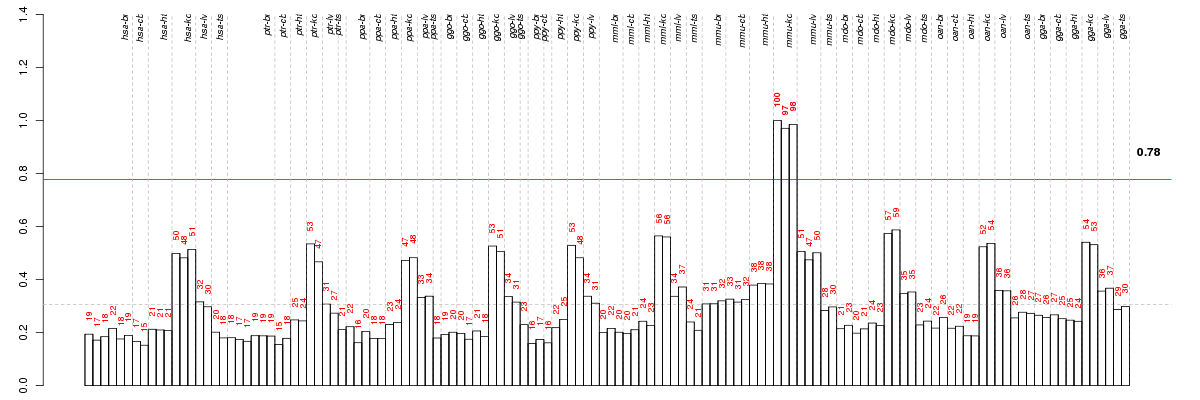

Under-expression is coded with green,
over-expression with red color.

transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
transmembrane transport
The process whereby a solute is transported from one side of a membrane to the other. This process includes the actual movement of the solute, and any regulation and preparatory steps, such as reduction of the solute.
all
NA
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
transmembrane transport
The process whereby a solute is transported from one side of a membrane to the other. This process includes the actual movement of the solute, and any regulation and preparatory steps, such as reduction of the solute.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
brush border
Dense covering of microvilli on the apical surface of epithelial cells in tissues such as the intestine, kidney, and choroid plexus; the microvilli aid absorption by increasing the surface area of the cell.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
brush border membrane
The portion of the plasma membrane surrounding the brush border.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
apical part of cell
The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
brush border membrane
The portion of the plasma membrane surrounding the brush border.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
secondary active transmembrane transporter activity
Catalysis of the transfer of a solute from one side of a membrane to the other, up its concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction and is driven by a chemiosmotic source of energy. Chemiosmotic sources of energy include uniport, symport or antiport.
symporter activity
Enables the active transport of a solute across a membrane by a mechanism whereby two or more species are transported together in the same direction in a tightly coupled process not directly linked to a form of energy other than chemiosmotic energy.
active transmembrane transporter activity
Catalysis of the transfer of a specific substance or related group of substances from one side of a membrane to the other, up the solute's concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04142 | 1.931e-02 | 3.445 | 11 | 51 | Lysosome |
| 00601 | 2.577e-02 | 0.4729 | 4 | 7 | Glycosphingolipid biosynthesis - lacto and neolacto series |
A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 1 ABCC2ATP-binding cassette, sub-family C (CFTR/MRP), member 2 (ENSG00000023839), score: 0.28 ABCC4ATP-binding cassette, sub-family C (CFTR/MRP), member 4 (ENSG00000125257), score: 0.35 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.36 ACO1aconitase 1, soluble (ENSG00000122729), score: 0.3 ACOT12acyl-CoA thioesterase 12 (ENSG00000172497), score: 0.31 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (ENSG00000049192), score: 0.28 ADAP2ArfGAP with dual PH domains 2 (ENSG00000184060), score: 0.31 AFTPHaftiphilin (ENSG00000119844), score: 0.29 AGPHD1aminoglycoside phosphotransferase domain containing 1 (ENSG00000188266), score: 0.38 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (ENSG00000117448), score: 0.37 ALDH8A1aldehyde dehydrogenase 8 family, member A1 (ENSG00000118514), score: 0.28 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.32 ANGEL1angel homolog 1 (Drosophila) (ENSG00000013523), score: -0.29 ANGEL2angel homolog 2 (Drosophila) (ENSG00000174606), score: 0.3 ANGPTL7angiopoietin-like 7 (ENSG00000171819), score: 0.28 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.48 ANKS4Bankyrin repeat and sterile alpha motif domain containing 4B (ENSG00000175311), score: 0.29 AOAHacyloxyacyl hydrolase (neutrophil) (ENSG00000136250), score: 0.54 AP1G1adaptor-related protein complex 1, gamma 1 subunit (ENSG00000166747), score: 0.31 AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.35 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.35 ARHGAP24Rho GTPase activating protein 24 (ENSG00000138639), score: 0.38 ARHGAP42Rho GTPase activating protein 42 (ENSG00000165895), score: 0.32 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 0.38 ARMC7armadillo repeat containing 7 (ENSG00000125449), score: -0.3 ARSBarylsulfatase B (ENSG00000113273), score: 0.46 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.29 ATP11AATPase, class VI, type 11A (ENSG00000068650), score: 0.47 ATP6V0A2ATPase, H+ transporting, lysosomal V0 subunit a2 (ENSG00000185344), score: 0.27 ATP6V0A4ATPase, H+ transporting, lysosomal V0 subunit a4 (ENSG00000105929), score: 0.37 ATP7AATPase, Cu++ transporting, alpha polypeptide (ENSG00000165240), score: 0.29 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: -0.31 ATXN7ataxin 7 (ENSG00000163635), score: 0.27 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.44 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (ENSG00000170340), score: 0.31 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.45 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: 0.28 BMP15bone morphogenetic protein 15 (ENSG00000130385), score: 0.37 BNC2basonuclin 2 (ENSG00000173068), score: 0.4 BPHLbiphenyl hydrolase-like (serine hydrolase) (ENSG00000137274), score: 0.29 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.58 C11orf54chromosome 11 open reading frame 54 (ENSG00000182919), score: 0.39 C12orf23chromosome 12 open reading frame 23 (ENSG00000151135), score: -0.29 C16orf68chromosome 16 open reading frame 68 (ENSG00000067365), score: -0.3 C16orf80chromosome 16 open reading frame 80 (ENSG00000070761), score: -0.36 C1orf100chromosome 1 open reading frame 100 (ENSG00000173728), score: 0.32 C1orf128chromosome 1 open reading frame 128 (ENSG00000057757), score: -0.33 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.32 C3orf75chromosome 3 open reading frame 75 (ENSG00000163832), score: -0.29 C5orf51chromosome 5 open reading frame 51 (ENSG00000205765), score: 0.31 C7orf57chromosome 7 open reading frame 57 (ENSG00000164746), score: 0.3 CA12carbonic anhydrase XII (ENSG00000074410), score: 0.29 CACHD1cache domain containing 1 (ENSG00000158966), score: 0.3 CALML4calmodulin-like 4 (ENSG00000129007), score: 0.34 CASP8caspase 8, apoptosis-related cysteine peptidase (ENSG00000064012), score: 0.32 CASRcalcium-sensing receptor (ENSG00000036828), score: 0.39 CCDC6coiled-coil domain containing 6 (ENSG00000108091), score: 0.33 CCDC90Acoiled-coil domain containing 90A (ENSG00000050393), score: 0.32 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.39 CDCP1CUB domain containing protein 1 (ENSG00000163814), score: 0.44 CDH2cadherin 2, type 1, N-cadherin (neuronal) (ENSG00000170558), score: -0.35 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.37 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: 0.37 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.39 CLDN16claudin 16 (ENSG00000113946), score: 0.37 CLDN19claudin 19 (ENSG00000164007), score: 0.31 CMTM6CKLF-like MARVEL transmembrane domain containing 6 (ENSG00000091317), score: 0.3 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.55 COL4A3collagen, type IV, alpha 3 (Goodpasture antigen) (ENSG00000169031), score: 0.41 COL4A4collagen, type IV, alpha 4 (ENSG00000081052), score: 0.45 CROTcarnitine O-octanoyltransferase (ENSG00000005469), score: 0.36 CRY1cryptochrome 1 (photolyase-like) (ENSG00000008405), score: -0.33 CRY2cryptochrome 2 (photolyase-like) (ENSG00000121671), score: -0.31 CUBNcubilin (intrinsic factor-cobalamin receptor) (ENSG00000107611), score: 0.38 CUX1cut-like homeobox 1 (ENSG00000160967), score: 0.29 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: 0.29 CYBRD1cytochrome b reductase 1 (ENSG00000071967), score: -0.32 DAB2disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) (ENSG00000153071), score: 0.31 DAOD-amino-acid oxidase (ENSG00000110887), score: 0.27 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.28 DCLK3doublecortin-like kinase 3 (ENSG00000163673), score: 0.35 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: -0.35 DKK2dickkopf homolog 2 (Xenopus laevis) (ENSG00000155011), score: 0.41 DMAP1DNA methyltransferase 1 associated protein 1 (ENSG00000178028), score: -0.3 DMXL1Dmx-like 1 (ENSG00000172869), score: 0.34 DNAJC12DnaJ (Hsp40) homolog, subfamily C, member 12 (ENSG00000108176), score: 0.34 DNAJC22DnaJ (Hsp40) homolog, subfamily C, member 22 (ENSG00000178401), score: 0.29 DYNC2LI1dynein, cytoplasmic 2, light intermediate chain 1 (ENSG00000138036), score: 0.29 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.27 EGFepidermal growth factor (ENSG00000138798), score: 0.53 EGFL6EGF-like-domain, multiple 6 (ENSG00000198759), score: 0.52 EHFets homologous factor (ENSG00000135373), score: 0.38 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (ENSG00000172071), score: 0.28 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.39 ERBB2IPerbb2 interacting protein (ENSG00000112851), score: 0.39 ERLIN2ER lipid raft associated 2 (ENSG00000147475), score: 0.32 ESRP1epithelial splicing regulatory protein 1 (ENSG00000104413), score: 0.38 EVCEllis van Creveld syndrome (ENSG00000072840), score: 0.39 EVI5ecotropic viral integration site 5 (ENSG00000067208), score: 0.31 EXOC6exocyst complex component 6 (ENSG00000138190), score: 0.38 F2RL1coagulation factor II (thrombin) receptor-like 1 (ENSG00000164251), score: 0.32 FAM116Bfamily with sequence similarity 116, member B (ENSG00000205593), score: -0.3 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: 0.28 FAM20Bfamily with sequence similarity 20, member B (ENSG00000116199), score: 0.28 FAR1fatty acyl CoA reductase 1 (ENSG00000197601), score: 0.36 FBP2fructose-1,6-bisphosphatase 2 (ENSG00000130957), score: 0.39 FBXO31F-box protein 31 (ENSG00000103264), score: -0.4 FCHO2FCH domain only 2 (ENSG00000157107), score: 0.39 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.36 FEZ1fasciculation and elongation protein zeta 1 (zygin I) (ENSG00000149557), score: -0.31 FGFBP1fibroblast growth factor binding protein 1 (ENSG00000137440), score: 0.43 FOXI1forkhead box I1 (ENSG00000168269), score: 0.3 FRRS1ferric-chelate reductase 1 (ENSG00000156869), score: 0.36 FUCA2fucosidase, alpha-L- 2, plasma (ENSG00000001036), score: 0.44 FUT9fucosyltransferase 9 (alpha (1,3) fucosyltransferase) (ENSG00000172461), score: 0.41 FYNFYN oncogene related to SRC, FGR, YES (ENSG00000010810), score: -0.28 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (ENSG00000141429), score: 0.35 GALNT11UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) (ENSG00000178234), score: 0.38 GALNT14UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) (ENSG00000158089), score: 0.3 GAS2growth arrest-specific 2 (ENSG00000148935), score: 0.43 GBA2glucosidase, beta (bile acid) 2 (ENSG00000070610), score: -0.45 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.31 GCNT1glucosaminyl (N-acetyl) transferase 1, core 2 (ENSG00000187210), score: 0.67 GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: 0.27 GIT2G protein-coupled receptor kinase interacting ArfGAP 2 (ENSG00000139436), score: -0.29 GJB2gap junction protein, beta 2, 26kDa (ENSG00000165474), score: 0.3 GLB1galactosidase, beta 1 (ENSG00000170266), score: 0.32 GM2AGM2 ganglioside activator (ENSG00000196743), score: 0.35 GNA13guanine nucleotide binding protein (G protein), alpha 13 (ENSG00000120063), score: 0.31 GRPEL2GrpE-like 2, mitochondrial (E. coli) (ENSG00000164284), score: 0.29 H1FXH1 histone family, member X (ENSG00000184897), score: -0.51 H6PDhexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (ENSG00000049239), score: 0.29 HAO2hydroxyacid oxidase 2 (long chain) (ENSG00000116882), score: 0.3 HECTD2HECT domain containing 2 (ENSG00000165338), score: -0.3 HELBhelicase (DNA) B (ENSG00000127311), score: 0.31 HEY1hairy/enhancer-of-split related with YRPW motif 1 (ENSG00000164683), score: -0.32 HINFPhistone H4 transcription factor (ENSG00000172273), score: -0.32 HNF4Ahepatocyte nuclear factor 4, alpha (ENSG00000101076), score: 0.29 HOXA10homeobox A10 (ENSG00000153807), score: 0.4 HOXB7homeobox B7 (ENSG00000120087), score: 0.39 IAH1isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) (ENSG00000134330), score: 0.28 IGF1Rinsulin-like growth factor 1 receptor (ENSG00000140443), score: 0.3 IGHMBP2immunoglobulin mu binding protein 2 (ENSG00000132740), score: -0.31 IL15interleukin 15 (ENSG00000164136), score: 0.3 IL5RAinterleukin 5 receptor, alpha (ENSG00000091181), score: 0.46 ILDR1immunoglobulin-like domain containing receptor 1 (ENSG00000145103), score: 0.41 INPP5Binositol polyphosphate-5-phosphatase, 75kDa (ENSG00000204084), score: 0.39 ITCHitchy E3 ubiquitin protein ligase homolog (mouse) (ENSG00000078747), score: 0.31 ITGB6integrin, beta 6 (ENSG00000115221), score: 0.41 ITGB8integrin, beta 8 (ENSG00000105855), score: 0.3 ITM2Aintegral membrane protein 2A (ENSG00000078596), score: -0.3 ITM2Bintegral membrane protein 2B (ENSG00000136156), score: 0.29 IVNS1ABPinfluenza virus NS1A binding protein (ENSG00000116679), score: 0.27 JAK2Janus kinase 2 (ENSG00000096968), score: 0.27 KCNJ1potassium inwardly-rectifying channel, subfamily J, member 1 (ENSG00000151704), score: 0.4 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (ENSG00000153822), score: 0.38 KIAA0100KIAA0100 (ENSG00000007202), score: 0.32 KIAA1614KIAA1614 (ENSG00000135835), score: 0.36 KIAA2018KIAA2018 (ENSG00000176542), score: 0.33 KIDINS220kinase D-interacting substrate, 220kDa (ENSG00000134313), score: 0.3 KLklotho (ENSG00000133116), score: 0.44 KLHDC2kelch domain containing 2 (ENSG00000165516), score: -0.3 KLHDC7Akelch domain containing 7A (ENSG00000179023), score: 0.29 KRT80keratin 80 (ENSG00000167767), score: 0.6 LAD1ladinin 1 (ENSG00000159166), score: 0.32 LAMP2lysosomal-associated membrane protein 2 (ENSG00000005893), score: 0.34 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.27 LDHDlactate dehydrogenase D (ENSG00000166816), score: 0.34 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: 0.32 LMBRD2LMBR1 domain containing 2 (ENSG00000164187), score: 0.35 LPAR3lysophosphatidic acid receptor 3 (ENSG00000171517), score: 0.27 LRRC20leucine rich repeat containing 20 (ENSG00000172731), score: -0.32 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.45 LRSAM1leucine rich repeat and sterile alpha motif containing 1 (ENSG00000148356), score: -0.3 M6PRmannose-6-phosphate receptor (cation dependent) (ENSG00000003056), score: 0.3 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: 0.36 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.28 MARCH6membrane-associated ring finger (C3HC4) 6 (ENSG00000145495), score: 0.29 MCCC1methylcrotonoyl-CoA carboxylase 1 (alpha) (ENSG00000078070), score: 0.36 MCM3APminichromosome maintenance complex component 3 associated protein (ENSG00000160294), score: -0.33 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.39 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.42 MEP1Ameprin A, alpha (PABA peptide hydrolase) (ENSG00000112818), score: 0.96 MEP1Bmeprin A, beta (ENSG00000141434), score: 0.98 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.3 MIPOL1mirror-image polydactyly 1 (ENSG00000151338), score: 0.31 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.58 MMP15matrix metallopeptidase 15 (membrane-inserted) (ENSG00000102996), score: -0.31 MUTmethylmalonyl CoA mutase (ENSG00000146085), score: 0.28 MYO6myosin VI (ENSG00000196586), score: 0.41 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.37 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.38 NFAT5nuclear factor of activated T-cells 5, tonicity-responsive (ENSG00000102908), score: 0.31 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.46 NPNTnephronectin (ENSG00000168743), score: 0.35 NR1H4nuclear receptor subfamily 1, group H, member 4 (ENSG00000012504), score: 0.29 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.41 NUAK2NUAK family, SNF1-like kinase, 2 (ENSG00000163545), score: 0.39 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.54 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: 0.33 NUP85nucleoporin 85kDa (ENSG00000125450), score: -0.3 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (ENSG00000173559), score: 0.28 OLFM4olfactomedin 4 (ENSG00000102837), score: 0.51 OPTNoptineurin (ENSG00000123240), score: -0.4 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.39 OSBPL6oxysterol binding protein-like 6 (ENSG00000079156), score: 0.3 OSGIN2oxidative stress induced growth inhibitor family member 2 (ENSG00000164823), score: -0.29 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.3 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.32 PAHphenylalanine hydroxylase (ENSG00000171759), score: 0.3 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.27 PAPSS23'-phosphoadenosine 5'-phosphosulfate synthase 2 (ENSG00000198682), score: 0.32 PARP1poly (ADP-ribose) polymerase 1 (ENSG00000143799), score: -0.3 PAWRPRKC, apoptosis, WT1, regulator (ENSG00000177425), score: 0.28 PEMTphosphatidylethanolamine N-methyltransferase (ENSG00000133027), score: -0.36 PICALMphosphatidylinositol binding clathrin assembly protein (ENSG00000073921), score: 0.33 PIGQphosphatidylinositol glycan anchor biosynthesis, class Q (ENSG00000007541), score: 0.3 PKD2polycystic kidney disease 2 (autosomal dominant) (ENSG00000118762), score: 0.4 PLAUplasminogen activator, urokinase (ENSG00000122861), score: 0.45 PLCXD2phosphatidylinositol-specific phospholipase C, X domain containing 2 (ENSG00000144824), score: 0.28 PLEKHF2pleckstrin homology domain containing, family F (with FYVE domain) member 2 (ENSG00000175895), score: 0.27 PLEKHN1pleckstrin homology domain containing, family N member 1 (ENSG00000187583), score: 0.29 PLS1plastin 1 (ENSG00000120756), score: 0.35 PLXDC2plexin domain containing 2 (ENSG00000120594), score: 0.38 POLR1Cpolymerase (RNA) I polypeptide C, 30kDa (ENSG00000171453), score: 0.28 PPA2pyrophosphatase (inorganic) 2 (ENSG00000138777), score: 0.28 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (ENSG00000132825), score: -0.28 PPP1R7protein phosphatase 1, regulatory (inhibitor) subunit 7 (ENSG00000115685), score: -0.35 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: -0.46 PQLC1PQ loop repeat containing 1 (ENSG00000122490), score: -0.29 PRKAG3protein kinase, AMP-activated, gamma 3 non-catalytic subunit (ENSG00000115592), score: 0.29 PRLRprolactin receptor (ENSG00000113494), score: 0.36 PROM1prominin 1 (ENSG00000007062), score: 0.36 PROSCproline synthetase co-transcribed homolog (bacterial) (ENSG00000147471), score: 0.3 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.3 PSMA4proteasome (prosome, macropain) subunit, alpha type, 4 (ENSG00000041357), score: 0.27 PTERphosphotriesterase related (ENSG00000165983), score: 0.38 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: 0.28 PYROXD2pyridine nucleotide-disulphide oxidoreductase domain 2 (ENSG00000119943), score: 0.27 RAB11FIP3RAB11 family interacting protein 3 (class II) (ENSG00000090565), score: 0.3 RAB38RAB38, member RAS oncogene family (ENSG00000123892), score: 0.35 RAB3IPRAB3A interacting protein (rabin3) (ENSG00000127328), score: 0.34 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.33 RBM9RNA binding motif protein 9 (ENSG00000100320), score: -0.3 RHCGRh family, C glycoprotein (ENSG00000140519), score: 0.27 RHOBTB3Rho-related BTB domain containing 3 (ENSG00000164292), score: -0.32 RNF141ring finger protein 141 (ENSG00000110315), score: -0.3 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.28 RRHretinal pigment epithelium-derived rhodopsin homolog (ENSG00000180245), score: 0.45 RRP12ribosomal RNA processing 12 homolog (S. cerevisiae) (ENSG00000052749), score: -0.28 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (ENSG00000211456), score: 0.29 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.29 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: -0.3 SCINscinderin (ENSG00000006747), score: 0.29 SCNN1Bsodium channel, nonvoltage-gated 1, beta (ENSG00000168447), score: 0.3 SEC62SEC62 homolog (S. cerevisiae) (ENSG00000008952), score: 0.27 SEP1515 kDa selenoprotein (ENSG00000183291), score: 0.33 SERINC3serine incorporator 3 (ENSG00000132824), score: 0.33 SESTD1SEC14 and spectrin domains 1 (ENSG00000187231), score: 0.33 SFMBT2Scm-like with four mbt domains 2 (ENSG00000198879), score: -0.33 SFXN1sideroflexin 1 (ENSG00000164466), score: 0.28 SGK2serum/glucocorticoid regulated kinase 2 (ENSG00000101049), score: 0.29 SGPP1sphingosine-1-phosphate phosphatase 1 (ENSG00000126821), score: 0.37 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: -0.35 SH2D4BSH2 domain containing 4B (ENSG00000178217), score: 0.29 SIM2single-minded homolog 2 (Drosophila) (ENSG00000159263), score: 0.43 SKAP2src kinase associated phosphoprotein 2 (ENSG00000005020), score: 0.28 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.46 SLC13A1solute carrier family 13 (sodium/sulfate symporters), member 1 (ENSG00000081800), score: 0.45 SLC13A3solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 (ENSG00000158296), score: 0.27 SLC16A12solute carrier family 16, member 12 (monocarboxylic acid transporter 12) (ENSG00000152779), score: 0.32 SLC16A4solute carrier family 16, member 4 (monocarboxylic acid transporter 5) (ENSG00000168679), score: 0.29 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (ENSG00000119899), score: 0.28 SLC18A1solute carrier family 18 (vesicular monoamine), member 1 (ENSG00000036565), score: 0.75 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.4 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: 0.37 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (ENSG00000100372), score: 0.39 SLC34A1solute carrier family 34 (sodium phosphate), member 1 (ENSG00000131183), score: 0.39 SLC35B4solute carrier family 35, member B4 (ENSG00000205060), score: 0.34 SLC39A8solute carrier family 39 (zinc transporter), member 8 (ENSG00000138821), score: 0.32 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (ENSG00000138079), score: 0.29 SLC41A3solute carrier family 41, member 3 (ENSG00000114544), score: -0.3 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (ENSG00000080493), score: 0.3 SLC5A11solute carrier family 5 (sodium/glucose cotransporter), member 11 (ENSG00000158865), score: 0.28 SLC5A12solute carrier family 5 (sodium/glucose cotransporter), member 12 (ENSG00000148942), score: 0.38 SLC6A19solute carrier family 6 (neutral amino acid transporter), member 19 (ENSG00000174358), score: 0.34 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.34 SLC7A9solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 (ENSG00000021488), score: 0.35 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.4 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.4 SNX11sorting nexin 11 (ENSG00000002919), score: -0.3 SNX29sorting nexin 29 (ENSG00000048471), score: 0.41 SNX6sorting nexin 6 (ENSG00000129515), score: 0.41 SNX8sorting nexin 8 (ENSG00000106266), score: 0.27 SOSTDC1sclerostin domain containing 1 (ENSG00000171243), score: 0.44 SOX9SRY (sex determining region Y)-box 9 (ENSG00000125398), score: -0.35 SPC25SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000152253), score: 0.34 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.34 SPSB4splA/ryanodine receptor domain and SOCS box containing 4 (ENSG00000175093), score: 0.31 SPTLC2serine palmitoyltransferase, long chain base subunit 2 (ENSG00000100596), score: 0.36 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.32 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (ENSG00000136840), score: -0.32 ST6GALNAC6ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 (ENSG00000160408), score: -0.36 STARD5StAR-related lipid transfer (START) domain containing 5 (ENSG00000172345), score: 0.3 STX7syntaxin 7 (ENSG00000079950), score: 0.3 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.29 SUSD2sushi domain containing 2 (ENSG00000099994), score: 0.38 SUSD3sushi domain containing 3 (ENSG00000157303), score: 0.27 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (ENSG00000204634), score: -0.35 TBCELtubulin folding cofactor E-like (ENSG00000154114), score: 0.33 TCN2transcobalamin II (ENSG00000185339), score: 0.39 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.27 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (ENSG00000100726), score: -0.31 TFAP2Btranscription factor AP-2 beta (activating enhancer binding protein 2 beta) (ENSG00000008196), score: 0.35 TFCP2L1transcription factor CP2-like 1 (ENSG00000115112), score: 0.38 TGFBR1transforming growth factor, beta receptor 1 (ENSG00000106799), score: 0.27 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (ENSG00000135966), score: 0.32 THNSL2threonine synthase-like 2 (S. cerevisiae) (ENSG00000144115), score: 0.27 THOC5THO complex 5 (ENSG00000100296), score: -0.28 TIMP3TIMP metallopeptidase inhibitor 3 (ENSG00000100234), score: 0.32 TINAGtubulointerstitial nephritis antigen (ENSG00000137251), score: 0.31 TM2D2TM2 domain containing 2 (ENSG00000169490), score: 0.31 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: 0.42 TMED6transmembrane emp24 protein transport domain containing 6 (ENSG00000157315), score: 0.36 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.34 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.3 TMEM14Atransmembrane protein 14A (ENSG00000096092), score: -0.3 TMEM20transmembrane protein 20 (ENSG00000176273), score: 0.31 TMEM27transmembrane protein 27 (ENSG00000147003), score: 0.42 TMEM64transmembrane protein 64 (ENSG00000180694), score: 0.4 TMIGD1transmembrane and immunoglobulin domain containing 1 (ENSG00000182271), score: 0.56 TMPRSS2transmembrane protease, serine 2 (ENSG00000184012), score: 0.33 TMTC2transmembrane and tetratricopeptide repeat containing 2 (ENSG00000179104), score: -0.28 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (ENSG00000145779), score: 0.55 TNIKTRAF2 and NCK interacting kinase (ENSG00000154310), score: -0.33 TNNtenascin N (ENSG00000120332), score: 0.32 TRIM7tripartite motif-containing 7 (ENSG00000146054), score: 0.36 TRPM6transient receptor potential cation channel, subfamily M, member 6 (ENSG00000119121), score: 0.32 TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: 0.31 TRPV4transient receptor potential cation channel, subfamily V, member 4 (ENSG00000111199), score: 0.37 TSHRthyroid stimulating hormone receptor (ENSG00000165409), score: 0.3 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: 0.29 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (ENSG00000197355), score: 0.32 UBLCP1ubiquitin-like domain containing CTD phosphatase 1 (ENSG00000164332), score: 0.27 UBTD2ubiquitin domain containing 2 (ENSG00000168246), score: -0.29 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.28 UPK3Auroplakin 3A (ENSG00000100373), score: 0.54 VASH1vasohibin 1 (ENSG00000071246), score: -0.35 VEZTvezatin, adherens junctions transmembrane protein (ENSG00000028203), score: -0.31 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: 0.31 VWA2von Willebrand factor A domain containing 2 (ENSG00000165816), score: 0.38 WDR36WD repeat domain 36 (ENSG00000134987), score: 0.29 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.3 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.4 WNT5Awingless-type MMTV integration site family, member 5A (ENSG00000114251), score: 0.27 XPNPEP1X-prolyl aminopeptidase (aminopeptidase P) 1, soluble (ENSG00000108039), score: 0.39 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.44 ZC3H12Czinc finger CCCH-type containing 12C (ENSG00000149289), score: 0.42 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.31 ZC3HC1zinc finger, C3HC-type containing 1 (ENSG00000091732), score: -0.28 ZCCHC11zinc finger, CCHC domain containing 11 (ENSG00000134744), score: -0.32 ZMYND19zinc finger, MYND-type containing 19 (ENSG00000165724), score: -0.34 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.39 ZNHIT3zinc finger, HIT type 3 (ENSG00000108278), score: -0.29 ZPLD1zona pellucida-like domain containing 1 (ENSG00000170044), score: 0.47
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |