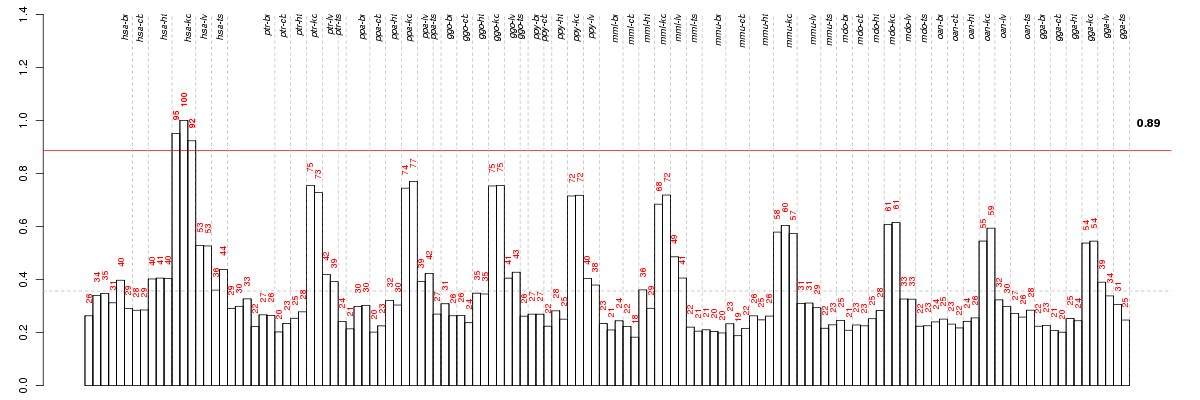

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
secretion
The controlled release of a substance by a cell, a group of cells, or a tissue.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
NA
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
sodium channel complex
An ion channel complex through which sodium ions pass.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
apical part of cell
The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
peptidase activity
Catalysis of the hydrolysis of a peptide bond. A peptide bond is a covalent bond formed when the carbon atom from the carboxyl group of one amino acid shares electrons with the nitrogen atom from the amino group of a second amino acid.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
secondary active transmembrane transporter activity
Catalysis of the transfer of a solute from one side of a membrane to the other, up its concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction and is driven by a chemiosmotic source of energy. Chemiosmotic sources of energy include uniport, symport or antiport.
exopeptidase activity
Catalysis of the hydrolysis of a peptide bond not more than three residues from the N- or C-terminus of a polypeptide chain, in a reaction that requires a free N-terminal amino group, C-terminal carboxyl group or both.
peptidase activity, acting on L-amino acid peptides
Catalysis of the hydrolysis of peptide bonds formed between L-amino acids.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
active transmembrane transporter activity
Catalysis of the transfer of a specific substance or related group of substances from one side of a membrane to the other, up the solute's concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction.
all
NA
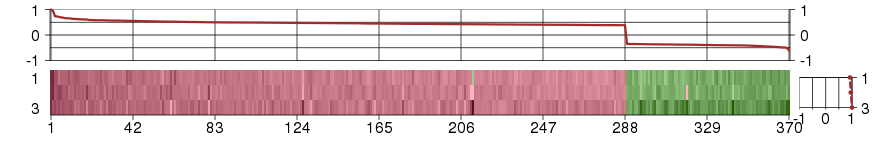
AATFapoptosis antagonizing transcription factor (ENSG00000108270), score: 0.41 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: -0.41 ACE2angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 (ENSG00000130234), score: 0.45 ACMSDaminocarboxymuconate semialdehyde decarboxylase (ENSG00000153086), score: 0.39 ACO1aconitase 1, soluble (ENSG00000122729), score: 0.41 ACOT11acyl-CoA thioesterase 11 (ENSG00000162390), score: 0.55 ACPPacid phosphatase, prostate (ENSG00000014257), score: 0.71 ACTN4actinin, alpha 4 (ENSG00000130402), score: 0.41 ADMadrenomedullin (ENSG00000148926), score: 0.39 ADPRHL2ADP-ribosylhydrolase like 2 (ENSG00000116863), score: 0.42 AGRPagouti related protein homolog (mouse) (ENSG00000159723), score: 0.42 AIM1absent in melanoma 1 (ENSG00000112297), score: 0.39 AIMP2aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 (ENSG00000106305), score: 0.4 ALS2CR4amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4 (ENSG00000155755), score: -0.36 ANKRA2ankyrin repeat, family A (RFXANK-like), 2 (ENSG00000164331), score: -0.36 ANO10anoctamin 10 (ENSG00000160746), score: 0.43 ANXA13annexin A13 (ENSG00000104537), score: 0.48 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: 0.44 APPBP2amyloid beta precursor protein (cytoplasmic tail) binding protein 2 (ENSG00000062725), score: -0.38 AQP3aquaporin 3 (Gill blood group) (ENSG00000165272), score: 0.56 ATF2activating transcription factor 2 (ENSG00000115966), score: -0.37 ATG10ATG10 autophagy related 10 homolog (S. cerevisiae) (ENSG00000152348), score: 0.46 ATP6V0A4ATPase, H+ transporting, lysosomal V0 subunit a4 (ENSG00000105929), score: 0.49 ATP6V0D2ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 (ENSG00000147614), score: 0.47 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (ENSG00000113732), score: 0.42 BACE2beta-site APP-cleaving enzyme 2 (ENSG00000182240), score: 0.49 BARX2BARX homeobox 2 (ENSG00000043039), score: 0.56 BBOX1butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 (ENSG00000129151), score: 0.48 BCAP29B-cell receptor-associated protein 29 (ENSG00000075790), score: -0.41 BDKRB1bradykinin receptor B1 (ENSG00000100739), score: 0.53 BDKRB2bradykinin receptor B2 (ENSG00000168398), score: 0.47 BNC2basonuclin 2 (ENSG00000173068), score: 0.42 C10orf137chromosome 10 open reading frame 137 (ENSG00000107938), score: -0.43 C10orf18chromosome 10 open reading frame 18 (ENSG00000108021), score: -0.35 C11orf54chromosome 11 open reading frame 54 (ENSG00000182919), score: 0.41 C14orf45chromosome 14 open reading frame 45 (ENSG00000119636), score: 0.54 C18orf1chromosome 18 open reading frame 1 (ENSG00000168675), score: -0.48 C19orf28chromosome 19 open reading frame 28 (ENSG00000161091), score: 0.53 C1orf107chromosome 1 open reading frame 107 (ENSG00000117597), score: -0.39 C22orf28chromosome 22 open reading frame 28 (ENSG00000100220), score: 0.54 C2orf18chromosome 2 open reading frame 18 (ENSG00000213699), score: 0.44 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.48 C3orf58chromosome 3 open reading frame 58 (ENSG00000181744), score: -0.4 C4orf31chromosome 4 open reading frame 31 (ENSG00000173376), score: 0.49 C9orf72chromosome 9 open reading frame 72 (ENSG00000147894), score: -0.46 CA12carbonic anhydrase XII (ENSG00000074410), score: 0.58 CAPN13calpain 13 (ENSG00000162949), score: 0.44 CASRcalcium-sensing receptor (ENSG00000036828), score: 0.48 CCNT2cyclin T2 (ENSG00000082258), score: -0.61 CD151CD151 molecule (Raph blood group) (ENSG00000177697), score: 0.4 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (ENSG00000113361), score: 0.46 CDK13cyclin-dependent kinase 13 (ENSG00000065883), score: -0.37 CELcarboxyl ester lipase (bile salt-stimulated lipase) (ENSG00000170835), score: 0.55 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: 0.48 CIITAclass II, major histocompatibility complex, transactivator (ENSG00000179583), score: 0.5 CISD2CDGSH iron sulfur domain 2 (ENSG00000145354), score: 0.43 CLCN7chloride channel 7 (ENSG00000103249), score: 0.43 CLDN16claudin 16 (ENSG00000113946), score: 0.45 CLDN19claudin 19 (ENSG00000164007), score: 0.45 CLN5ceroid-lipofuscinosis, neuronal 5 (ENSG00000102805), score: 0.41 CLN6ceroid-lipofuscinosis, neuronal 6, late infantile, variant (ENSG00000128973), score: 0.49 CLPTM1LCLPTM1-like (ENSG00000049656), score: 0.49 COQ4coenzyme Q4 homolog (S. cerevisiae) (ENSG00000167113), score: 0.47 COX15COX15 homolog, cytochrome c oxidase assembly protein (yeast) (ENSG00000014919), score: 0.42 CPA1carboxypeptidase A1 (pancreatic) (ENSG00000091704), score: 0.42 CPEB3cytoplasmic polyadenylation element binding protein 3 (ENSG00000107864), score: -0.43 CPMcarboxypeptidase M (ENSG00000135678), score: 0.39 CRABP1cellular retinoic acid binding protein 1 (ENSG00000166426), score: 0.42 CRTAPcartilage associated protein (ENSG00000170275), score: 0.43 CRYAAcrystallin, alpha A (ENSG00000160202), score: 0.74 CRYL1crystallin, lambda 1 (ENSG00000165475), score: 0.49 CTNND1catenin (cadherin-associated protein), delta 1 (ENSG00000198561), score: 0.41 CTSHcathepsin H (ENSG00000103811), score: 0.52 CTTNcortactin (ENSG00000085733), score: 0.46 CUBNcubilin (intrinsic factor-cobalamin receptor) (ENSG00000107611), score: 0.52 CXCL14chemokine (C-X-C motif) ligand 14 (ENSG00000145824), score: 0.44 D2HGDHD-2-hydroxyglutarate dehydrogenase (ENSG00000180902), score: 0.42 DAAM1dishevelled associated activator of morphogenesis 1 (ENSG00000100592), score: -0.49 DAB2disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) (ENSG00000153071), score: 0.46 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.43 DDCdopa decarboxylase (aromatic L-amino acid decarboxylase) (ENSG00000132437), score: 0.53 DDHD1DDHD domain containing 1 (ENSG00000100523), score: -0.41 DENND2DDENN/MADD domain containing 2D (ENSG00000162777), score: 0.47 DENND5BDENN/MADD domain containing 5B (ENSG00000170456), score: -0.42 DEPDC6DEP domain containing 6 (ENSG00000155792), score: 0.46 DGKEdiacylglycerol kinase, epsilon 64kDa (ENSG00000153933), score: -0.35 DLEC1deleted in lung and esophageal cancer 1 (ENSG00000008226), score: 0.49 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: -0.37 DOK4docking protein 4 (ENSG00000125170), score: 0.42 DPEP1dipeptidase 1 (renal) (ENSG00000015413), score: 0.57 DPH1DPH1 homolog (S. cerevisiae) (ENSG00000108963), score: 0.48 DPP4dipeptidyl-peptidase 4 (ENSG00000197635), score: 0.43 DUSP10dual specificity phosphatase 10 (ENSG00000143507), score: -0.42 EEA1early endosome antigen 1 (ENSG00000102189), score: -0.38 EGFepidermal growth factor (ENSG00000138798), score: 0.4 EHFets homologous factor (ENSG00000135373), score: 0.4 ELF3E74-like factor 3 (ets domain transcription factor, epithelial-specific ) (ENSG00000163435), score: 0.49 ELF5E74-like factor 5 (ets domain transcription factor) (ENSG00000135374), score: 0.58 ELMO3engulfment and cell motility 3 (ENSG00000102890), score: 0.44 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.46 ENPEPglutamyl aminopeptidase (aminopeptidase A) (ENSG00000138792), score: 0.41 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.46 EPC2enhancer of polycomb homolog 2 (Drosophila) (ENSG00000135999), score: -0.39 EPCAMepithelial cell adhesion molecule (ENSG00000119888), score: 0.44 ERBB2v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) (ENSG00000141736), score: 0.45 ERGIC1endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 (ENSG00000113719), score: 0.39 ERGIC2ERGIC and golgi 2 (ENSG00000087502), score: -0.38 ESRP1epithelial splicing regulatory protein 1 (ENSG00000104413), score: 0.47 EXOC3exocyst complex component 3 (ENSG00000180104), score: 0.51 EXOC6exocyst complex component 6 (ENSG00000138190), score: -0.36 EXOSC7exosome component 7 (ENSG00000075914), score: 0.5 F2RL1coagulation factor II (thrombin) receptor-like 1 (ENSG00000164251), score: 0.49 FAM150Bfamily with sequence similarity 150, member B (ENSG00000189292), score: 0.69 FAM160B1family with sequence similarity 160, member B1 (ENSG00000151553), score: -0.38 FAM3Bfamily with sequence similarity 3, member B (ENSG00000183844), score: 0.45 FAM55Cfamily with sequence similarity 55, member C (ENSG00000144815), score: -0.39 FAM76Bfamily with sequence similarity 76, member B (ENSG00000077458), score: -0.38 FAM83Hfamily with sequence similarity 83, member H (ENSG00000180921), score: 0.47 FARP1FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) (ENSG00000152767), score: 0.43 FBXL20F-box and leucine-rich repeat protein 20 (ENSG00000108306), score: -0.4 FBXL5F-box and leucine-rich repeat protein 5 (ENSG00000118564), score: 0.54 FBXO11F-box protein 11 (ENSG00000138081), score: -0.39 FBXO33F-box protein 33 (ENSG00000165355), score: -0.4 FBXO7F-box protein 7 (ENSG00000100225), score: 0.41 FBXW7F-box and WD repeat domain containing 7 (ENSG00000109670), score: -0.39 FGD5FYVE, RhoGEF and PH domain containing 5 (ENSG00000154783), score: 0.42 FGFR4fibroblast growth factor receptor 4 (ENSG00000160867), score: 0.42 FLOT2flotillin 2 (ENSG00000132589), score: 0.39 FOXI1forkhead box I1 (ENSG00000168269), score: 0.55 FRMD6FERM domain containing 6 (ENSG00000139926), score: -0.46 FUCA2fucosidase, alpha-L- 2, plasma (ENSG00000001036), score: 0.44 GALNT14UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) (ENSG00000158089), score: 0.49 GANgigaxonin (ENSG00000127688), score: -0.36 GATA3GATA binding protein 3 (ENSG00000107485), score: 0.51 GCNT3glucosaminyl (N-acetyl) transferase 3, mucin type (ENSG00000140297), score: 0.58 GLB1galactosidase, beta 1 (ENSG00000170266), score: 0.44 GLG1golgi glycoprotein 1 (ENSG00000090863), score: 0.5 GOPCgolgi-associated PDZ and coiled-coil motif containing (ENSG00000047932), score: -0.38 GORABgolgin, RAB6-interacting (ENSG00000120370), score: -0.36 GPCPD1glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) (ENSG00000125772), score: -0.4 GPR114G protein-coupled receptor 114 (ENSG00000159618), score: 1 GPR132G protein-coupled receptor 132 (ENSG00000183484), score: 0.44 GPR56G protein-coupled receptor 56 (ENSG00000205336), score: 0.48 GRHL2grainyhead-like 2 (Drosophila) (ENSG00000083307), score: 0.48 HAO2hydroxyacid oxidase 2 (long chain) (ENSG00000116882), score: 0.39 HDHD3haloacid dehalogenase-like hydrolase domain containing 3 (ENSG00000119431), score: 0.4 HECTD2HECT domain containing 2 (ENSG00000165338), score: -0.4 HELZhelicase with zinc finger (ENSG00000198265), score: -0.37 HEMK1HemK methyltransferase family member 1 (ENSG00000114735), score: 0.43 HMBOX1homeobox containing 1 (ENSG00000147421), score: -0.35 HNF1AHNF1 homeobox A (ENSG00000135100), score: 0.45 HOOK3hook homolog 3 (Drosophila) (ENSG00000168172), score: -0.41 HOXA10homeobox A10 (ENSG00000153807), score: 0.48 HOXB7homeobox B7 (ENSG00000120087), score: 0.41 HOXD10homeobox D10 (ENSG00000128710), score: 0.46 HOXD4homeobox D4 (ENSG00000170166), score: 0.49 HSD11B2hydroxysteroid (11-beta) dehydrogenase 2 (ENSG00000176387), score: 0.51 HYAL2hyaluronoglucosaminidase 2 (ENSG00000068001), score: 0.39 ICMTisoprenylcysteine carboxyl methyltransferase (ENSG00000116237), score: 0.49 IGF2BP1insulin-like growth factor 2 mRNA binding protein 1 (ENSG00000159217), score: 0.62 IL1RL1interleukin 1 receptor-like 1 (ENSG00000115602), score: 0.61 IL22RA1interleukin 22 receptor, alpha 1 (ENSG00000142677), score: 0.53 ILDR1immunoglobulin-like domain containing receptor 1 (ENSG00000145103), score: 0.51 IMPA2inositol(myo)-1(or 4)-monophosphatase 2 (ENSG00000141401), score: 0.54 INTS2integrator complex subunit 2 (ENSG00000108506), score: -0.49 INTS8integrator complex subunit 8 (ENSG00000164941), score: -0.38 INVSinversin (ENSG00000119509), score: 0.41 ITM2Bintegral membrane protein 2B (ENSG00000136156), score: 0.47 KCNE3potassium voltage-gated channel, Isk-related family, member 3 (ENSG00000175538), score: 0.64 KCNJ1potassium inwardly-rectifying channel, subfamily J, member 1 (ENSG00000151704), score: 0.56 KCNK5potassium channel, subfamily K, member 5 (ENSG00000164626), score: 0.53 KCNQ1potassium voltage-gated channel, KQT-like subfamily, member 1 (ENSG00000053918), score: 0.44 KIAA0100KIAA0100 (ENSG00000007202), score: 0.4 KIAA0391KIAA0391 (ENSG00000100890), score: 0.52 KIAA1370KIAA1370 (ENSG00000047346), score: -0.36 KIAA1522KIAA1522 (ENSG00000162522), score: 0.39 KIF12kinesin family member 12 (ENSG00000136883), score: 0.42 KIF13Bkinesin family member 13B (ENSG00000197892), score: 0.52 KLklotho (ENSG00000133116), score: 0.46 KLHDC7Akelch domain containing 7A (ENSG00000179023), score: 0.43 KLHL28kelch-like 28 (Drosophila) (ENSG00000179454), score: -0.4 LAD1ladinin 1 (ENSG00000159166), score: 0.39 LGALS2lectin, galactoside-binding, soluble, 2 (ENSG00000100079), score: 0.55 LLGL2lethal giant larvae homolog 2 (Drosophila) (ENSG00000073350), score: 0.49 LMX1BLIM homeobox transcription factor 1, beta (ENSG00000136944), score: 0.66 LOC652522similar to Werner syndrome protein (ENSG00000165392), score: -0.38 LPIN3lipin 3 (ENSG00000132793), score: 0.41 LRRC8Cleucine rich repeat containing 8 family, member C (ENSG00000171488), score: -0.4 MALmal, T-cell differentiation protein (ENSG00000172005), score: 0.44 MALLmal, T-cell differentiation protein-like (ENSG00000144063), score: 0.41 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: 0.39 MED13mediator complex subunit 13 (ENSG00000108510), score: -0.38 METTL6methyltransferase like 6 (ENSG00000206562), score: -0.4 METTL9methyltransferase like 9 (ENSG00000197006), score: 0.44 MMABmethylmalonic aciduria (cobalamin deficiency) cblB type (ENSG00000139428), score: 0.4 MMP7matrix metallopeptidase 7 (matrilysin, uterine) (ENSG00000137673), score: 0.62 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: 0.45 MSNmoesin (ENSG00000147065), score: 0.43 MSRAmethionine sulfoxide reductase A (ENSG00000175806), score: 0.39 MYO1Dmyosin ID (ENSG00000176658), score: 0.43 MYOM3myomesin family, member 3 (ENSG00000142661), score: 0.51 NAA25N(alpha)-acetyltransferase 25, NatB auxiliary subunit (ENSG00000111300), score: -0.44 NDRG4NDRG family member 4 (ENSG00000103034), score: -0.35 NEDD9neural precursor cell expressed, developmentally down-regulated 9 (ENSG00000111859), score: 0.4 NINJ1ninjurin 1 (ENSG00000131669), score: 0.44 NME7non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000143156), score: 0.41 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.58 NPHS2nephrosis 2, idiopathic, steroid-resistant (podocin) (ENSG00000116218), score: 0.61 NPLOC4nuclear protein localization 4 homolog (S. cerevisiae) (ENSG00000182446), score: 0.46 NPNTnephronectin (ENSG00000168743), score: 0.45 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (ENSG00000113389), score: 0.43 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (ENSG00000107960), score: 0.39 OGDHLoxoglutarate dehydrogenase-like (ENSG00000197444), score: 0.44 OGG18-oxoguanine DNA glycosylase (ENSG00000114026), score: 0.47 OLFM4olfactomedin 4 (ENSG00000102837), score: 0.56 ORC6Lorigin recognition complex, subunit 6 like (yeast) (ENSG00000091651), score: -0.37 OVCH2ovochymase 2 (gene/pseudogene) (ENSG00000183378), score: 0.49 P4HA2prolyl 4-hydroxylase, alpha polypeptide II (ENSG00000072682), score: 0.42 PABPC1Lpoly(A) binding protein, cytoplasmic 1-like (ENSG00000101104), score: 0.56 PAPPApregnancy-associated plasma protein A, pappalysin 1 (ENSG00000182752), score: 0.52 PAPPA2pappalysin 2 (ENSG00000116183), score: 0.95 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: -0.35 PCGF2polycomb group ring finger 2 (ENSG00000056661), score: 0.4 PDFpeptide deformylase (mitochondrial) (ENSG00000213380), score: 0.45 PDS5BPDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) (ENSG00000083642), score: -0.38 PELI1pellino homolog 1 (Drosophila) (ENSG00000197329), score: -0.41 PEPDpeptidase D (ENSG00000124299), score: 0.63 PFKLphosphofructokinase, liver (ENSG00000141959), score: 0.47 PIGQphosphatidylinositol glycan anchor biosynthesis, class Q (ENSG00000007541), score: 0.47 PLA2G15phospholipase A2, group XV (ENSG00000103066), score: 0.5 PLA2R1phospholipase A2 receptor 1, 180kDa (ENSG00000153246), score: 0.44 PLATplasminogen activator, tissue (ENSG00000104368), score: 0.48 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (ENSG00000197943), score: 0.49 PLEK2pleckstrin 2 (ENSG00000100558), score: 0.41 PLEKHJ1pleckstrin homology domain containing, family J member 1 (ENSG00000104886), score: 0.66 PNPLA1patatin-like phospholipase domain containing 1 (ENSG00000180316), score: 0.47 POFUT1protein O-fucosyltransferase 1 (ENSG00000101346), score: 0.39 POLR2Cpolymerase (RNA) II (DNA directed) polypeptide C, 33kDa (ENSG00000102978), score: 0.4 POLR2Hpolymerase (RNA) II (DNA directed) polypeptide H (ENSG00000163882), score: 0.44 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: 0.46 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (ENSG00000111725), score: 0.56 PROM2prominin 2 (ENSG00000155066), score: 0.57 PROX1prospero homeobox 1 (ENSG00000117707), score: -0.37 PRRG4proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) (ENSG00000135378), score: 0.41 PSKH1protein serine kinase H1 (ENSG00000159792), score: 0.42 PSMD9proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 (ENSG00000110801), score: 0.55 PSTKphosphoseryl-tRNA kinase (ENSG00000179988), score: -0.37 PTBP2polypyrimidine tract binding protein 2 (ENSG00000117569), score: -0.39 PTPN1protein tyrosine phosphatase, non-receptor type 1 (ENSG00000196396), score: 0.52 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: 0.57 RAB11FIP1RAB11 family interacting protein 1 (class I) (ENSG00000156675), score: 0.4 RAB11FIP3RAB11 family interacting protein 3 (class II) (ENSG00000090565), score: 0.47 RAB17RAB17, member RAS oncogene family (ENSG00000124839), score: 0.55 RAB33BRAB33B, member RAS oncogene family (ENSG00000172007), score: -0.45 RAB38RAB38, member RAS oncogene family (ENSG00000123892), score: 0.39 RAB3GAP2RAB3 GTPase activating protein subunit 2 (non-catalytic) (ENSG00000118873), score: -0.36 RAB7L1RAB7, member RAS oncogene family-like 1 (ENSG00000117280), score: 0.45 RAMP3receptor (G protein-coupled) activity modifying protein 3 (ENSG00000122679), score: 0.4 RASL11ARAS-like, family 11, member A (ENSG00000122035), score: 0.39 RB1CC1RB1-inducible coiled-coil 1 (ENSG00000023287), score: -0.5 RBM26RNA binding motif protein 26 (ENSG00000139746), score: -0.45 RFXANKregulatory factor X-associated ankyrin-containing protein (ENSG00000064490), score: 0.43 RHCGRh family, C glycoprotein (ENSG00000140519), score: 0.55 RICTORRPTOR independent companion of MTOR, complex 2 (ENSG00000164327), score: -0.41 RIPK4receptor-interacting serine-threonine kinase 4 (ENSG00000183421), score: 0.41 RLFrearranged L-myc fusion (ENSG00000117000), score: -0.38 RNF152ring finger protein 152 (ENSG00000176641), score: 0.47 RSPO1R-spondin homolog (Xenopus laevis) (ENSG00000169218), score: 0.45 SCINscinderin (ENSG00000006747), score: 0.47 SCNN1Asodium channel, nonvoltage-gated 1 alpha (ENSG00000111319), score: 0.54 SCNN1Bsodium channel, nonvoltage-gated 1, beta (ENSG00000168447), score: 0.59 SCNN1Gsodium channel, nonvoltage-gated 1, gamma (ENSG00000166828), score: 0.53 SCRN2secernin 2 (ENSG00000141295), score: 0.49 SDF4stromal cell derived factor 4 (ENSG00000078808), score: 0.41 SEC13SEC13 homolog (S. cerevisiae) (ENSG00000157020), score: 0.39 SFXN2sideroflexin 2 (ENSG00000156398), score: 0.45 SGK196protein kinase-like protein SgK196 (ENSG00000185900), score: -0.45 SGK2serum/glucocorticoid regulated kinase 2 (ENSG00000101049), score: 0.4 SIM2single-minded homolog 2 (Drosophila) (ENSG00000159263), score: 0.51 SLC13A1solute carrier family 13 (sodium/sulfate symporters), member 1 (ENSG00000081800), score: 0.43 SLC13A3solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 (ENSG00000158296), score: 0.47 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (ENSG00000088386), score: 0.73 SLC16A4solute carrier family 16, member 4 (monocarboxylic acid transporter 5) (ENSG00000168679), score: 0.45 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (ENSG00000119899), score: 0.39 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.47 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.47 SLC22A7solute carrier family 22 (organic anion transporter), member 7 (ENSG00000137204), score: 0.39 SLC25A42solute carrier family 25, member 42 (ENSG00000181035), score: 0.49 SLC26A9solute carrier family 26, member 9 (ENSG00000174502), score: 0.47 SLC2A9solute carrier family 2 (facilitated glucose transporter), member 9 (ENSG00000109667), score: 0.43 SLC34A1solute carrier family 34 (sodium phosphate), member 1 (ENSG00000131183), score: 0.48 SLC39A13solute carrier family 39 (zinc transporter), member 13 (ENSG00000165915), score: 0.5 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (ENSG00000138079), score: 0.44 SLC44A3solute carrier family 44, member 3 (ENSG00000143036), score: 0.48 SLC5A12solute carrier family 5 (sodium/glucose cotransporter), member 12 (ENSG00000148942), score: 0.57 SLC5A9solute carrier family 5 (sodium/glucose cotransporter), member 9 (ENSG00000117834), score: 0.48 SLC6A19solute carrier family 6 (neutral amino acid transporter), member 19 (ENSG00000174358), score: 0.59 SLC7A9solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 (ENSG00000021488), score: 0.53 SLC9A3solute carrier family 9 (sodium/hydrogen exchanger), member 3 (ENSG00000066230), score: 0.5 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (ENSG00000109062), score: 0.4 SLC9A4solute carrier family 9 (sodium/hydrogen exchanger), member 4 (ENSG00000180251), score: 0.66 SNX13sorting nexin 13 (ENSG00000071189), score: -0.38 SNX22sorting nexin 22 (ENSG00000157734), score: 0.51 SNX29sorting nexin 29 (ENSG00000048471), score: 0.39 SNX5sorting nexin 5 (ENSG00000089006), score: 0.44 SOX13SRY (sex determining region Y)-box 13 (ENSG00000143842), score: 0.4 SPASTspastin (ENSG00000021574), score: -0.41 SPNS2spinster homolog 2 (Drosophila) (ENSG00000183018), score: 0.39 SPP1secreted phosphoprotein 1 (ENSG00000118785), score: 0.48 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (ENSG00000172296), score: 0.52 SRSF11serine/arginine-rich splicing factor 11 (ENSG00000116754), score: -0.4 ST14suppression of tumorigenicity 14 (colon carcinoma) (ENSG00000149418), score: 0.49 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (ENSG00000115525), score: -0.37 STAP1signal transducing adaptor family member 1 (ENSG00000035720), score: 0.4 STC1stanniocalcin 1 (ENSG00000159167), score: 0.42 STK32Bserine/threonine kinase 32B (ENSG00000152953), score: 0.5 STXBP5syntaxin binding protein 5 (tomosyn) (ENSG00000164506), score: -0.43 SUCLG1succinate-CoA ligase, alpha subunit (ENSG00000163541), score: 0.49 SULF2sulfatase 2 (ENSG00000196562), score: 0.41 SUMF1sulfatase modifying factor 1 (ENSG00000144455), score: 0.43 SUSD1sushi domain containing 1 (ENSG00000106868), score: 0.47 SUSD2sushi domain containing 2 (ENSG00000099994), score: 0.61 SUSD3sushi domain containing 3 (ENSG00000157303), score: 0.5 TBC1D1TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 (ENSG00000065882), score: 0.51 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (ENSG00000204634), score: 0.41 TBX2T-box 2 (ENSG00000121068), score: 0.49 TFCP2L1transcription factor CP2-like 1 (ENSG00000115112), score: 0.58 TH1LTH1-like (Drosophila) (ENSG00000101158), score: 0.58 TINAGtubulointerstitial nephritis antigen (ENSG00000137251), score: 0.45 TJP3tight junction protein 3 (zona occludens 3) (ENSG00000105289), score: 0.41 TM7SF3transmembrane 7 superfamily member 3 (ENSG00000064115), score: 0.57 TMCO4transmembrane and coiled-coil domains 4 (ENSG00000162542), score: 0.47 TMEM100transmembrane protein 100 (ENSG00000166292), score: -0.35 TMEM171transmembrane protein 171 (ENSG00000157111), score: 0.47 TMEM27transmembrane protein 27 (ENSG00000147003), score: 0.4 TMEM51transmembrane protein 51 (ENSG00000171729), score: 0.42 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.49 TMPRSS2transmembrane protease, serine 2 (ENSG00000184012), score: 0.49 TMPRSS3transmembrane protease, serine 3 (ENSG00000160183), score: 0.41 TMPRSS4transmembrane protease, serine 4 (ENSG00000137648), score: 0.68 TNRC6Ctrinucleotide repeat containing 6C (ENSG00000078687), score: -0.47 TPP2tripeptidyl peptidase II (ENSG00000134900), score: -0.37 TRIM23tripartite motif-containing 23 (ENSG00000113595), score: -0.4 TRPV4transient receptor potential cation channel, subfamily V, member 4 (ENSG00000111199), score: 0.52 TULP3tubby like protein 3 (ENSG00000078246), score: 0.4 UFSP2UFM1-specific peptidase 2 (ENSG00000109775), score: 0.46 UNC45Aunc-45 homolog A (C. elegans) (ENSG00000140553), score: 0.46 VCAM1vascular cell adhesion molecule 1 (ENSG00000162692), score: 0.42 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (ENSG00000175073), score: -0.4 VGLL4vestigial like 4 (Drosophila) (ENSG00000144560), score: 0.4 VRK3vaccinia related kinase 3 (ENSG00000105053), score: 0.43 WARS2tryptophanyl tRNA synthetase 2, mitochondrial (ENSG00000116874), score: 0.47 WBP1WW domain binding protein 1 (ENSG00000115274), score: 0.39 WDR1WD repeat domain 1 (ENSG00000071127), score: 0.54 WDR34WD repeat domain 34 (ENSG00000119333), score: 0.45 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.63 WDR91WD repeat domain 91 (ENSG00000105875), score: 0.57 WNT8Bwingless-type MMTV integration site family, member 8B (ENSG00000075290), score: 0.5 WWP2WW domain containing E3 ubiquitin protein ligase 2 (ENSG00000198373), score: 0.41 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.61 YAF2YY1 associated factor 2 (ENSG00000015153), score: -0.37 ZBTB46zinc finger and BTB domain containing 46 (ENSG00000130584), score: 0.39 ZBTB6zinc finger and BTB domain containing 6 (ENSG00000186130), score: -0.49 ZCCHC11zinc finger, CCHC domain containing 11 (ENSG00000134744), score: -0.37 ZDHHC20zinc finger, DHHC-type containing 20 (ENSG00000180776), score: -0.43 ZFATzinc finger and AT hook domain containing (ENSG00000066827), score: 0.51 ZGPATzinc finger, CCCH-type with G patch domain (ENSG00000197114), score: 0.39 ZNF292zinc finger protein 292 (ENSG00000188994), score: -0.39 ZNF518Bzinc finger protein 518B (ENSG00000178163), score: -0.42 ZNF644zinc finger protein 644 (ENSG00000122482), score: -0.47
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |