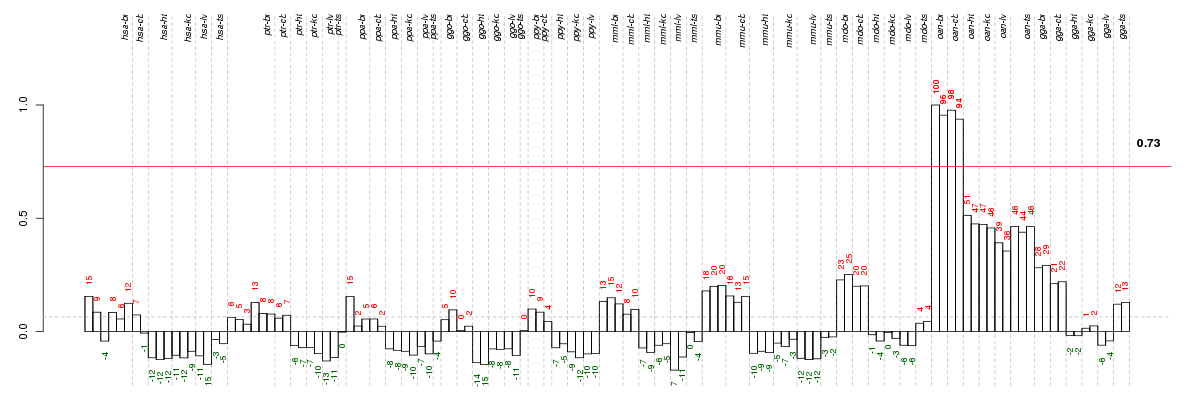

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04620 | 3.190e-03 | 1.828 | 9 | 30 | Toll-like receptor signaling pathway |
| 04210 | 1.788e-02 | 1.888 | 8 | 31 | Apoptosis |
| 04722 | 2.947e-02 | 3.107 | 10 | 51 | Neurotrophin signaling pathway |
| 05220 | 4.927e-02 | 1.34 | 6 | 22 | Chronic myeloid leukemia |
AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.68 AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: 0.43 ACSF3acyl-CoA synthetase family member 3 (ENSG00000176715), score: -0.32 ACTN4actinin, alpha 4 (ENSG00000130402), score: -0.38 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: -0.38 ADSSL1adenylosuccinate synthase like 1 (ENSG00000185100), score: -0.35 AFF4AF4/FMR2 family, member 4 (ENSG00000072364), score: -0.37 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (ENSG00000135744), score: -0.32 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: -0.5 AMZ1archaelysin family metallopeptidase 1 (ENSG00000174945), score: 0.44 ANKRA2ankyrin repeat, family A (RFXANK-like), 2 (ENSG00000164331), score: 0.36 ANKRD27ankyrin repeat domain 27 (VPS9 domain) (ENSG00000105186), score: -0.38 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: -0.45 AP4E1adaptor-related protein complex 4, epsilon 1 subunit (ENSG00000081014), score: 0.47 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: -0.36 AQRaquarius homolog (mouse) (ENSG00000021776), score: 0.43 ARHGAP44Rho GTPase activating protein 44 (ENSG00000006740), score: 0.36 ARHGEF18Rho/Rac guanine nucleotide exchange factor (GEF) 18 (ENSG00000104880), score: -0.33 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (ENSG00000163947), score: -0.39 ARIH2ariadne homolog 2 (Drosophila) (ENSG00000177479), score: -0.46 ARR3arrestin 3, retinal (X-arrestin) (ENSG00000120500), score: 0.43 ATG13ATG13 autophagy related 13 homolog (S. cerevisiae) (ENSG00000175224), score: 0.37 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.39 ATRXalpha thalassemia/mental retardation syndrome X-linked (ENSG00000085224), score: 0.39 ATXN7ataxin 7 (ENSG00000163635), score: -0.44 AVPR1Barginine vasopressin receptor 1B (ENSG00000198049), score: 1 AZI15-azacytidine induced 1 (ENSG00000141577), score: -0.44 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.37 BCDIN3DBCDIN3 domain containing (ENSG00000186666), score: -0.4 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.4 BMP3bone morphogenetic protein 3 (ENSG00000152785), score: 0.53 C10orf28chromosome 10 open reading frame 28 (ENSG00000166024), score: -0.35 C12orf29chromosome 12 open reading frame 29 (ENSG00000133641), score: 0.47 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.51 C14orf126chromosome 14 open reading frame 126 (ENSG00000129480), score: 0.38 C14orf37chromosome 14 open reading frame 37 (ENSG00000139971), score: 0.5 C15orf41chromosome 15 open reading frame 41 (ENSG00000186073), score: 0.38 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: -0.45 C1orf107chromosome 1 open reading frame 107 (ENSG00000117597), score: 0.46 C1orf25chromosome 1 open reading frame 25 (ENSG00000121486), score: 0.6 C20orf4chromosome 20 open reading frame 4 (ENSG00000131043), score: 0.47 C5orf33chromosome 5 open reading frame 33 (ENSG00000152620), score: -0.33 C6orf106chromosome 6 open reading frame 106 (ENSG00000196821), score: -0.42 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: 0.58 C7orf27chromosome 7 open reading frame 27 (ENSG00000106009), score: -0.34 C8orf40chromosome 8 open reading frame 40 (ENSG00000176209), score: -0.46 C9orf102chromosome 9 open reading frame 102 (ENSG00000182150), score: 0.39 CACNG2calcium channel, voltage-dependent, gamma subunit 2 (ENSG00000166862), score: 0.39 CAND2cullin-associated and neddylation-dissociated 2 (putative) (ENSG00000144712), score: -0.33 CAPN2calpain 2, (m/II) large subunit (ENSG00000162909), score: -0.36 CASD1CAS1 domain containing 1 (ENSG00000127995), score: 0.37 CASTcalpastatin (ENSG00000153113), score: -0.34 CBFBcore-binding factor, beta subunit (ENSG00000067955), score: -0.34 CCDC109Acoiled-coil domain containing 109A (ENSG00000156026), score: 0.48 CCDC127coiled-coil domain containing 127 (ENSG00000164366), score: -0.41 CCDC93coiled-coil domain containing 93 (ENSG00000125633), score: 0.46 CCNL1cyclin L1 (ENSG00000163660), score: -0.34 CDC37L1cell division cycle 37 homolog (S. cerevisiae)-like 1 (ENSG00000106993), score: -0.37 CDC40cell division cycle 40 homolog (S. cerevisiae) (ENSG00000168438), score: 0.46 CDC45cell division cycle 45 homolog (S. cerevisiae) (ENSG00000093009), score: 0.4 CDH12cadherin 12, type 2 (N-cadherin 2) (ENSG00000154162), score: 0.38 CDH20cadherin 20, type 2 (ENSG00000101542), score: 0.38 CDK15cyclin-dependent kinase 15 (ENSG00000138395), score: 0.43 CDK17cyclin-dependent kinase 17 (ENSG00000059758), score: 0.38 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.51 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.41 CENPHcentromere protein H (ENSG00000153044), score: 0.4 CENPQcentromere protein Q (ENSG00000031691), score: 0.35 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.46 CHRNB4cholinergic receptor, nicotinic, beta 4 (ENSG00000117971), score: 0.57 CHRNDcholinergic receptor, nicotinic, delta (ENSG00000135902), score: 0.36 CLCN3chloride channel 3 (ENSG00000109572), score: 0.52 CLIP1CAP-GLY domain containing linker protein 1 (ENSG00000130779), score: -0.33 CLK4CDC-like kinase 4 (ENSG00000113240), score: -0.37 CLPTM1LCLPTM1-like (ENSG00000049656), score: -0.32 CLSPNclaspin (ENSG00000092853), score: 0.39 CNTN5contactin 5 (ENSG00000149972), score: 0.72 COG2component of oligomeric golgi complex 2 (ENSG00000135775), score: 0.36 COL25A1collagen, type XXV, alpha 1 (ENSG00000188517), score: 0.47 COMMD2COMM domain containing 2 (ENSG00000114744), score: 0.43 COMMD8COMM domain containing 8 (ENSG00000169019), score: 0.41 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -0.57 CPLX4complexin 4 (ENSG00000166569), score: 0.75 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.41 CRB1crumbs homolog 1 (Drosophila) (ENSG00000134376), score: 0.35 CRISPLD1cysteine-rich secretory protein LCCL domain containing 1 (ENSG00000121005), score: 0.45 CRKv-crk sarcoma virus CT10 oncogene homolog (avian) (ENSG00000167193), score: -0.44 CRYABcrystallin, alpha B (ENSG00000109846), score: -0.33 CTHRC1collagen triple helix repeat containing 1 (ENSG00000164932), score: 0.47 CTNND1catenin (cadherin-associated protein), delta 1 (ENSG00000198561), score: -0.43 CUL4Acullin 4A (ENSG00000139842), score: -0.41 DAZAP1DAZ associated protein 1 (ENSG00000071626), score: -0.37 DCNdecorin (ENSG00000011465), score: -0.41 DCTN4dynactin 4 (p62) (ENSG00000132912), score: 0.6 DFFADNA fragmentation factor, 45kDa, alpha polypeptide (ENSG00000160049), score: 0.48 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.38 DIRC2disrupted in renal carcinoma 2 (ENSG00000138463), score: 0.36 DIS3LDIS3 mitotic control homolog (S. cerevisiae)-like (ENSG00000166938), score: -0.4 DKK2dickkopf homolog 2 (Xenopus laevis) (ENSG00000155011), score: 0.48 DPYSL3dihydropyrimidinase-like 3 (ENSG00000113657), score: 0.4 DRD2dopamine receptor D2 (ENSG00000149295), score: 0.38 DSCC1defective in sister chromatid cohesion 1 homolog (S. cerevisiae) (ENSG00000136982), score: 0.48 DSELdermatan sulfate epimerase-like (ENSG00000171451), score: 0.42 DUSP19dual specificity phosphatase 19 (ENSG00000162999), score: 0.4 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.41 EIF2B4eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa (ENSG00000115211), score: -0.33 EIF2C2eukaryotic translation initiation factor 2C, 2 (ENSG00000123908), score: -0.36 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (ENSG00000158711), score: -0.36 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.58 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.46 EPHA8EPH receptor A8 (ENSG00000070886), score: 0.42 EPHX4epoxide hydrolase 4 (ENSG00000172031), score: 0.39 EPS15epidermal growth factor receptor pathway substrate 15 (ENSG00000085832), score: 0.45 ERCC6Lexcision repair cross-complementing rodent repair deficiency, complementation group 6-like (ENSG00000186871), score: 0.37 ESR2estrogen receptor 2 (ER beta) (ENSG00000140009), score: 0.43 EXOC1exocyst complex component 1 (ENSG00000090989), score: 0.4 EXOC2exocyst complex component 2 (ENSG00000112685), score: 0.36 EXTL2exostoses (multiple)-like 2 (ENSG00000162694), score: 0.58 FADDFas (TNFRSF6)-associated via death domain (ENSG00000168040), score: -0.42 FAM164Afamily with sequence similarity 164, member A (ENSG00000104427), score: 0.52 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: -0.33 FAM185Afamily with sequence similarity 185, member A (ENSG00000222011), score: -0.44 FAM57Afamily with sequence similarity 57, member A (ENSG00000167695), score: 0.38 FAM63Bfamily with sequence similarity 63, member B (ENSG00000128923), score: 0.39 FAM69Afamily with sequence similarity 69, member A (ENSG00000154511), score: 0.4 FARP2FERM, RhoGEF and pleckstrin domain protein 2 (ENSG00000006607), score: -0.43 FBXO11F-box protein 11 (ENSG00000138081), score: 0.36 FBXO9F-box protein 9 (ENSG00000112146), score: 0.46 FCHO2FCH domain only 2 (ENSG00000157107), score: -0.33 FGF10fibroblast growth factor 10 (ENSG00000070193), score: 0.45 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.38 FIP1L1FIP1 like 1 (S. cerevisiae) (ENSG00000145216), score: 0.41 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: -0.32 FLNBfilamin B, beta (ENSG00000136068), score: -0.52 FOXP1forkhead box P1 (ENSG00000114861), score: -0.35 FRMPD4FERM and PDZ domain containing 4 (ENSG00000169933), score: 0.39 FUT11fucosyltransferase 11 (alpha (1,3) fucosyltransferase) (ENSG00000196968), score: -0.4 GAAglucosidase, alpha; acid (ENSG00000171298), score: -0.38 GABRA2gamma-aminobutyric acid (GABA) A receptor, alpha 2 (ENSG00000151834), score: 0.47 GABRA4gamma-aminobutyric acid (GABA) A receptor, alpha 4 (ENSG00000109158), score: 0.42 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (ENSG00000141429), score: 0.39 GALR1galanin receptor 1 (ENSG00000166573), score: 0.38 GLT25D1glycosyltransferase 25 domain containing 1 (ENSG00000130309), score: -0.34 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: -0.36 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (ENSG00000111670), score: -0.38 GOLGA1golgin A1 (ENSG00000136935), score: -0.42 GPBP1L1GC-rich promoter binding protein 1-like 1 (ENSG00000159592), score: -0.33 GPN3GPN-loop GTPase 3 (ENSG00000111231), score: -0.36 GPR12G protein-coupled receptor 12 (ENSG00000132975), score: 0.42 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 0.4 GPR149G protein-coupled receptor 149 (ENSG00000174948), score: 0.55 GPR85G protein-coupled receptor 85 (ENSG00000164604), score: 0.36 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.52 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.37 GUCY2Cguanylate cyclase 2C (heat stable enterotoxin receptor) (ENSG00000070019), score: 0.59 HEATR5BHEAT repeat containing 5B (ENSG00000008869), score: 0.4 HECW2HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 (ENSG00000138411), score: 0.45 HELQhelicase, POLQ-like (ENSG00000163312), score: 0.43 HERPUD1homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 (ENSG00000051108), score: -0.41 HHLA1HERV-H LTR-associating 1 (ENSG00000132297), score: 0.86 HMGXB3HMG box domain containing 3 (ENSG00000113716), score: 0.38 HNRNPRheterogeneous nuclear ribonucleoprotein R (ENSG00000125944), score: 0.43 HRH4histamine receptor H4 (ENSG00000134489), score: 0.49 HSF4heat shock transcription factor 4 (ENSG00000102878), score: -0.34 IAH1isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) (ENSG00000134330), score: -0.35 IBSPintegrin-binding sialoprotein (ENSG00000029559), score: 0.53 ICMTisoprenylcysteine carboxyl methyltransferase (ENSG00000116237), score: -0.38 IFT20intraflagellar transport 20 homolog (Chlamydomonas) (ENSG00000109083), score: -0.35 IKBKBinhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (ENSG00000104365), score: -0.35 IL12RB2interleukin 12 receptor, beta 2 (ENSG00000081985), score: 0.57 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.48 IL1RAPL2interleukin 1 receptor accessory protein-like 2 (ENSG00000189108), score: 0.44 IMPAD1inositol monophosphatase domain containing 1 (ENSG00000104331), score: 0.47 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: 0.49 KBTBD3kelch repeat and BTB (POZ) domain containing 3 (ENSG00000182359), score: 0.52 KCTD7potassium channel tetramerisation domain containing 7 (ENSG00000154710), score: 0.36 KIAA1279KIAA1279 (ENSG00000198954), score: 0.39 KLHDC2kelch domain containing 2 (ENSG00000165516), score: 0.39 KLHL8kelch-like 8 (Drosophila) (ENSG00000145332), score: 0.37 LANCL1LanC lantibiotic synthetase component C-like 1 (bacterial) (ENSG00000115365), score: 0.37 LAPTM5lysosomal protein transmembrane 5 (ENSG00000162511), score: -0.36 LARP4BLa ribonucleoprotein domain family, member 4B (ENSG00000107929), score: -0.37 LFNGLFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (ENSG00000106003), score: 0.36 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.45 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.38 LMNB2lamin B2 (ENSG00000176619), score: -0.33 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: -0.44 LOC100134291similar to mitogen-activated protein kinase phosphatase x (ENSG00000112679), score: -0.4 LOC100134387similar to DEAH (Asp-Glu-Ala-His) box polypeptide 40 (ENSG00000108406), score: -0.35 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.46 LOC100293905similar to gastrin-releasing peptide receptor (ENSG00000126010), score: 0.66 LOC653155PRP4 pre-mRNA processing factor 4 homolog B (yeast) pseudogene (ENSG00000112739), score: -0.39 LOH12CR1loss of heterozygosity, 12, chromosomal region 1 (ENSG00000165714), score: 0.42 LPXNleupaxin (ENSG00000110031), score: -0.34 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (ENSG00000126243), score: 0.38 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.69 LSAMPlimbic system-associated membrane protein (ENSG00000185565), score: 0.36 LXNlatexin (ENSG00000079257), score: 0.41 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: 0.44 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: 0.44 MAP2K1mitogen-activated protein kinase kinase 1 (ENSG00000169032), score: 0.37 MAPK8mitogen-activated protein kinase 8 (ENSG00000107643), score: 0.37 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (ENSG00000162889), score: -0.38 MBOAT1membrane bound O-acyltransferase domain containing 1 (ENSG00000172197), score: 0.41 MED23mediator complex subunit 23 (ENSG00000112282), score: 0.39 MEGF9multiple EGF-like-domains 9 (ENSG00000106780), score: 0.39 MIIPmigration and invasion inhibitory protein (ENSG00000116691), score: -0.32 MKLN1muskelin 1, intracellular mediator containing kelch motifs (ENSG00000128585), score: -0.33 MLXIPMLX interacting protein (ENSG00000175727), score: -0.52 MRPL12mitochondrial ribosomal protein L12 (ENSG00000183093), score: -0.49 MYO19myosin XIX (ENSG00000141140), score: -0.44 MYO9Bmyosin IXB (ENSG00000099331), score: -0.37 N6AMT2N-6 adenine-specific DNA methyltransferase 2 (putative) (ENSG00000150456), score: 0.45 NAIF1nuclear apoptosis inducing factor 1 (ENSG00000171169), score: 0.36 NECAP2NECAP endocytosis associated 2 (ENSG00000157191), score: -0.43 NFATC3nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 (ENSG00000072736), score: -0.4 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (ENSG00000109320), score: -0.48 NGBneuroglobin (ENSG00000165553), score: 0.45 NMNAT3nicotinamide nucleotide adenylyltransferase 3 (ENSG00000163864), score: -0.33 NPATnuclear protein, ataxia-telangiectasia locus (ENSG00000149308), score: 0.45 NPRL2nitrogen permease regulator-like 2 (S. cerevisiae) (ENSG00000114388), score: -0.43 NR2C2nuclear receptor subfamily 2, group C, member 2 (ENSG00000177463), score: -0.33 NT5DC25'-nucleotidase domain containing 2 (ENSG00000168268), score: -0.46 NTSneurotensin (ENSG00000133636), score: 0.38 NUDCD2NudC domain containing 2 (ENSG00000170584), score: 0.42 NUFIP2nuclear fragile X mental retardation protein interacting protein 2 (ENSG00000108256), score: -0.36 OFCC1orofacial cleft 1 candidate 1 (ENSG00000181355), score: 0.44 OLIG3oligodendrocyte transcription factor 3 (ENSG00000177468), score: 0.35 PACRGLPARK2 co-regulated-like (ENSG00000163138), score: 0.37 PAFAH1B2platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) (ENSG00000168092), score: 0.37 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: 0.4 PANX1pannexin 1 (ENSG00000110218), score: 0.45 PARVAparvin, alpha (ENSG00000197702), score: 0.37 PARVBparvin, beta (ENSG00000188677), score: -0.33 PCGF3polycomb group ring finger 3 (ENSG00000185619), score: -0.45 PCGF5polycomb group ring finger 5 (ENSG00000180628), score: -0.49 PDE5Aphosphodiesterase 5A, cGMP-specific (ENSG00000138735), score: 0.37 PDRG1p53 and DNA-damage regulated 1 (ENSG00000088356), score: 0.51 PFKFB46-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (ENSG00000114268), score: 0.38 PIGXphosphatidylinositol glycan anchor biosynthesis, class X (ENSG00000163964), score: 0.46 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (ENSG00000171608), score: 0.47 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (ENSG00000105647), score: -0.35 PLA2G15phospholipase A2, group XV (ENSG00000103066), score: -0.38 PNOCprepronociceptin (ENSG00000168081), score: 0.38 POC1APOC1 centriolar protein homolog A (Chlamydomonas) (ENSG00000164087), score: -0.32 POLGpolymerase (DNA directed), gamma (ENSG00000140521), score: -0.45 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (ENSG00000132664), score: 0.5 PPPDE2PPPDE peptidase domain containing 2 (ENSG00000100418), score: 0.42 PQLC3PQ loop repeat containing 3 (ENSG00000162976), score: -0.38 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (ENSG00000101161), score: -0.41 PRPH2peripherin 2 (retinal degeneration, slow) (ENSG00000112619), score: 0.58 PSKH1protein serine kinase H1 (ENSG00000159792), score: -0.32 PTP4A2protein tyrosine phosphatase type IVA, member 2 (ENSG00000184007), score: -0.43 PTPRAprotein tyrosine phosphatase, receptor type, A (ENSG00000132670), score: 0.54 PUS1pseudouridylate synthase 1 (ENSG00000177192), score: -0.41 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.51 RAB3GAP2RAB3 GTPase activating protein subunit 2 (non-catalytic) (ENSG00000118873), score: 0.44 RAB4ARAB4A, member RAS oncogene family (ENSG00000168118), score: 0.37 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.41 RBM45RNA binding motif protein 45 (ENSG00000155636), score: 0.37 RCHY1ring finger and CHY zinc finger domain containing 1 (ENSG00000163743), score: 0.36 RFC3replication factor C (activator 1) 3, 38kDa (ENSG00000133119), score: 0.47 RGMBRGM domain family, member B (ENSG00000174136), score: 0.37 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.44 RIC3resistance to inhibitors of cholinesterase 3 homolog (C. elegans) (ENSG00000166405), score: 0.4 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (ENSG00000137275), score: -0.36 RNF114ring finger protein 114 (ENSG00000124226), score: -0.45 RNF182ring finger protein 182 (ENSG00000180537), score: 0.38 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: 0.54 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: -0.56 RPE65retinal pigment epithelium-specific protein 65kDa (ENSG00000116745), score: 0.41 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (ENSG00000117676), score: -0.35 RPS6KC1ribosomal protein S6 kinase, 52kDa, polypeptide 1 (ENSG00000136643), score: 0.6 RRP9ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) (ENSG00000114767), score: -0.41 RXRAretinoid X receptor, alpha (ENSG00000186350), score: -0.44 SAP30LSAP30-like (ENSG00000164576), score: 0.38 SCGNsecretagogin, EF-hand calcium binding protein (ENSG00000079689), score: 0.39 SDCCAG8serologically defined colon cancer antigen 8 (ENSG00000054282), score: -0.34 SEMA3Esema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E (ENSG00000170381), score: 0.35 SEMA6Dsema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D (ENSG00000137872), score: 0.38 SERINC4serine incorporator 4 (ENSG00000184716), score: -0.33 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (ENSG00000197632), score: 0.46 SFRS18splicing factor, arginine/serine-rich 18 (ENSG00000132424), score: -0.34 SGPP1sphingosine-1-phosphate phosphatase 1 (ENSG00000126821), score: 0.36 SGTBsmall glutamine-rich tetratricopeptide repeat (TPR)-containing, beta (ENSG00000197860), score: 0.37 SH2B3SH2B adaptor protein 3 (ENSG00000111252), score: -0.44 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.36 SLC35A5solute carrier family 35, member A5 (ENSG00000138459), score: -0.54 SLC35F2solute carrier family 35, member F2 (ENSG00000110660), score: 0.38 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.48 SLC9A4solute carrier family 9 (sodium/hydrogen exchanger), member 4 (ENSG00000180251), score: 0.38 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: 0.39 SMC5structural maintenance of chromosomes 5 (ENSG00000198887), score: -0.38 SNX14sorting nexin 14 (ENSG00000135317), score: 0.38 SPASTspastin (ENSG00000021574), score: 0.46 SPATA5L1spermatogenesis associated 5-like 1 (ENSG00000171763), score: 0.41 SPENspen homolog, transcriptional regulator (Drosophila) (ENSG00000065526), score: -0.37 SPON1spondin 1, extracellular matrix protein (ENSG00000152268), score: 0.39 SRIsorcin (ENSG00000075142), score: 0.48 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: -0.39 STAB1stabilin 1 (ENSG00000010327), score: -0.38 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (ENSG00000124214), score: 0.46 STK40serine/threonine kinase 40 (ENSG00000196182), score: -0.33 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.54 STX12syntaxin 12 (ENSG00000117758), score: 0.46 TAPT1transmembrane anterior posterior transformation 1 (ENSG00000169762), score: -0.33 TBC1D19TBC1 domain family, member 19 (ENSG00000109680), score: 0.43 TBCEtubulin folding cofactor E (ENSG00000116957), score: 0.41 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: 0.4 TDRD7tudor domain containing 7 (ENSG00000196116), score: -0.47 TFAP2Dtranscription factor AP-2 delta (activating enhancer binding protein 2 delta) (ENSG00000008197), score: 0.37 THOC7THO complex 7 homolog (Drosophila) (ENSG00000163634), score: -0.52 THSD7Athrombospondin, type I, domain containing 7A (ENSG00000005108), score: 0.37 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.42 TMEM117transmembrane protein 117 (ENSG00000139173), score: 0.59 TMEM128transmembrane protein 128 (ENSG00000132406), score: 0.44 TMEM50Atransmembrane protein 50A (ENSG00000183726), score: 0.48 TNMDtenomodulin (ENSG00000000005), score: 0.37 TNRC6Atrinucleotide repeat containing 6A (ENSG00000090905), score: -0.38 TOM1L1target of myb1 (chicken)-like 1 (ENSG00000141198), score: -0.36 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: -0.48 TOR1Btorsin family 1, member B (torsin B) (ENSG00000136816), score: 0.38 TPBGtrophoblast glycoprotein (ENSG00000146242), score: 0.48 TPRA1transmembrane protein, adipocyte asscociated 1 (ENSG00000163870), score: -0.37 TRAF3TNF receptor-associated factor 3 (ENSG00000131323), score: -0.35 TRAPPC4trafficking protein particle complex 4 (ENSG00000196655), score: 0.41 TRDMT1tRNA aspartic acid methyltransferase 1 (ENSG00000107614), score: 0.36 TRIM23tripartite motif-containing 23 (ENSG00000113595), score: 0.39 TRIP13thyroid hormone receptor interactor 13 (ENSG00000071539), score: 0.38 TRIP4thyroid hormone receptor interactor 4 (ENSG00000103671), score: 0.36 TRNAU1APtRNA selenocysteine 1 associated protein 1 (ENSG00000180098), score: 0.41 TRPA1transient receptor potential cation channel, subfamily A, member 1 (ENSG00000104321), score: 0.55 TRPC5transient receptor potential cation channel, subfamily C, member 5 (ENSG00000072315), score: 0.46 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: -0.43 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.53 UBA6ubiquitin-like modifier activating enzyme 6 (ENSG00000033178), score: 0.37 UBL3ubiquitin-like 3 (ENSG00000122042), score: 0.45 UIMC1ubiquitin interaction motif containing 1 (ENSG00000087206), score: -0.36 UPF0639UPF0639 protein (ENSG00000175985), score: 0.37 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: -0.34 UTP15UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) (ENSG00000164338), score: 0.38 UTP23UTP23, small subunit (SSU) processome component, homolog (yeast) (ENSG00000147679), score: 0.38 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.67 VPS16vacuolar protein sorting 16 homolog (S. cerevisiae) (ENSG00000215305), score: 0.36 VPS4Bvacuolar protein sorting 4 homolog B (S. cerevisiae) (ENSG00000119541), score: 0.35 WACWW domain containing adaptor with coiled-coil (ENSG00000095787), score: -0.32 WDR26WD repeat domain 26 (ENSG00000162923), score: -0.46 WDR3WD repeat domain 3 (ENSG00000065183), score: 0.49 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.5 WDR76WD repeat domain 76 (ENSG00000092470), score: 0.37 WNT3Awingless-type MMTV integration site family, member 3A (ENSG00000154342), score: 0.9 WWP2WW domain containing E3 ubiquitin protein ligase 2 (ENSG00000198373), score: -0.37 XRCC4X-ray repair complementing defective repair in Chinese hamster cells 4 (ENSG00000152422), score: 0.38 YTHDC2YTH domain containing 2 (ENSG00000047188), score: -0.53 ZCCHC11zinc finger, CCHC domain containing 11 (ENSG00000134744), score: -0.36 ZFP161zinc finger protein 161 homolog (mouse) (ENSG00000198081), score: 0.4 ZHX1zinc fingers and homeoboxes 1 (ENSG00000165156), score: 0.43 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.53 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.33 ZW10ZW10, kinetochore associated, homolog (Drosophila) (ENSG00000086827), score: 0.37
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| oan_br_m_ca1 | oan | br | m | _ |