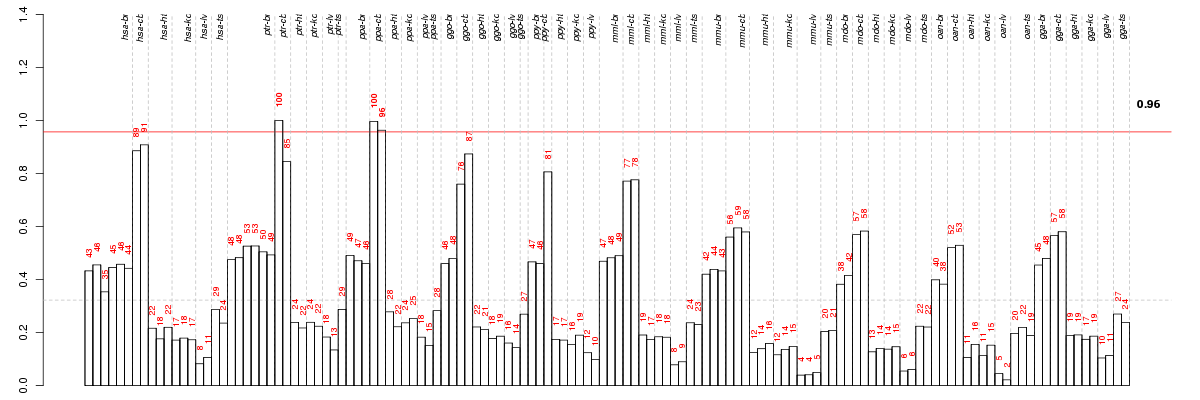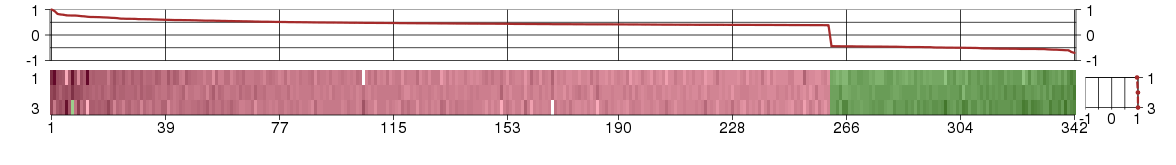

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.


AASDHaminoadipate-semialdehyde dehydrogenase (ENSG00000157426), score: 0.53 ABCC8ATP-binding cassette, sub-family C (CFTR/MRP), member 8 (ENSG00000006071), score: 0.43 ABI3ABI family, member 3 (ENSG00000108798), score: -0.46 ABLIM1actin binding LIM protein 1 (ENSG00000099204), score: 0.44 ACADSacyl-CoA dehydrogenase, C-2 to C-3 short chain (ENSG00000122971), score: -0.53 ACVR2Bactivin A receptor, type IIB (ENSG00000114739), score: 0.56 ADAM10ADAM metallopeptidase domain 10 (ENSG00000137845), score: 0.61 ADAM22ADAM metallopeptidase domain 22 (ENSG00000008277), score: 0.42 AFAP1L2actin filament associated protein 1-like 2 (ENSG00000169129), score: 0.45 AFF4AF4/FMR2 family, member 4 (ENSG00000072364), score: 0.4 AKAP12A kinase (PRKA) anchor protein 12 (ENSG00000131016), score: 0.39 AKAP13A kinase (PRKA) anchor protein 13 (ENSG00000170776), score: -0.54 ANKRD28ankyrin repeat domain 28 (ENSG00000206560), score: 0.62 ANKS6ankyrin repeat and sterile alpha motif domain containing 6 (ENSG00000165138), score: 0.49 AP4B1adaptor-related protein complex 4, beta 1 subunit (ENSG00000134262), score: 0.47 ARID4AAT rich interactive domain 4A (RBP1-like) (ENSG00000032219), score: 0.39 ARMC8armadillo repeat containing 8 (ENSG00000114098), score: 0.41 ARPC2actin related protein 2/3 complex, subunit 2, 34kDa (ENSG00000163466), score: -0.44 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (ENSG00000170340), score: -0.55 BAHCC1BAH domain and coiled-coil containing 1 (ENSG00000171282), score: 0.41 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.51 BPTFbromodomain PHD finger transcription factor (ENSG00000171634), score: 0.64 BTBD3BTB (POZ) domain containing 3 (ENSG00000132640), score: 0.49 C12orf51chromosome 12 open reading frame 51 (ENSG00000173064), score: 0.54 C14orf38chromosome 14 open reading frame 38 (ENSG00000151838), score: 0.5 C15orf17chromosome 15 open reading frame 17 (ENSG00000178761), score: 0.41 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.54 C16orf73chromosome 16 open reading frame 73 (ENSG00000162039), score: 0.46 C22orf28chromosome 22 open reading frame 28 (ENSG00000100220), score: -0.51 C6orf115chromosome 6 open reading frame 115 (ENSG00000146386), score: -0.67 C6orf57chromosome 6 open reading frame 57 (ENSG00000154079), score: -0.5 C7orf16chromosome 7 open reading frame 16 (ENSG00000106341), score: 0.48 C7orf55chromosome 7 open reading frame 55 (ENSG00000164898), score: -0.5 C8orf34chromosome 8 open reading frame 34 (ENSG00000165084), score: 0.56 C8orf79chromosome 8 open reading frame 79 (ENSG00000170941), score: 0.45 CA10carbonic anhydrase X (ENSG00000154975), score: 0.42 CACNB4calcium channel, voltage-dependent, beta 4 subunit (ENSG00000182389), score: 0.45 CAMKK2calcium/calmodulin-dependent protein kinase kinase 2, beta (ENSG00000110931), score: 0.45 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.62 CCDC109Acoiled-coil domain containing 109A (ENSG00000156026), score: 0.41 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: 0.4 CCDC82coiled-coil domain containing 82 (ENSG00000149231), score: 0.39 CCNJLcyclin J-like (ENSG00000135083), score: 0.71 CCNT2cyclin T2 (ENSG00000082258), score: 0.4 CDH18cadherin 18, type 2 (ENSG00000145526), score: 0.42 CDH23cadherin-related 23 (ENSG00000107736), score: 0.51 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.63 CDKN1Bcyclin-dependent kinase inhibitor 1B (p27, Kip1) (ENSG00000111276), score: 0.45 CDONCdon homolog (mouse) (ENSG00000064309), score: 0.58 CDR2Lcerebellar degeneration-related protein 2-like (ENSG00000109089), score: 0.4 CELcarboxyl ester lipase (bile salt-stimulated lipase) (ENSG00000170835), score: 0.54 CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: 0.55 CERKLceramide kinase-like (ENSG00000188452), score: 0.49 CHD9chromodomain helicase DNA binding protein 9 (ENSG00000177200), score: 0.39 CHGBchromogranin B (secretogranin 1) (ENSG00000089199), score: 0.46 CHN2chimerin (chimaerin) 2 (ENSG00000106069), score: 0.49 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.75 CLIC4chloride intracellular channel 4 (ENSG00000169504), score: -0.47 CLIP1CAP-GLY domain containing linker protein 1 (ENSG00000130779), score: 0.42 CLK4CDC-like kinase 4 (ENSG00000113240), score: 0.41 CLVS2clavesin 2 (ENSG00000146352), score: 0.46 CNOT2CCR4-NOT transcription complex, subunit 2 (ENSG00000111596), score: 0.58 CNTN6contactin 6 (ENSG00000134115), score: 0.7 CNTNAP1contactin associated protein 1 (ENSG00000108797), score: 0.39 COG3component of oligomeric golgi complex 3 (ENSG00000136152), score: 0.59 COL19A1collagen, type XIX, alpha 1 (ENSG00000082293), score: 0.63 COL7A1collagen, type VII, alpha 1 (ENSG00000114270), score: 0.47 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (ENSG00000173085), score: -0.44 CPEB4cytoplasmic polyadenylation element binding protein 4 (ENSG00000113742), score: 0.39 CPPED1calcineurin-like phosphoesterase domain containing 1 (ENSG00000103381), score: -0.51 CRAMP1LCrm, cramped-like (Drosophila) (ENSG00000007545), score: 0.53 CRHR1corticotropin releasing hormone receptor 1 (ENSG00000120088), score: 0.44 CRTAMcytotoxic and regulatory T cell molecule (ENSG00000109943), score: 0.83 CTCFCCCTC-binding factor (zinc finger protein) (ENSG00000102974), score: 0.42 CXCL12chemokine (C-X-C motif) ligand 12 (ENSG00000107562), score: -0.47 CYBASC3cytochrome b, ascorbate dependent 3 (ENSG00000162144), score: -0.47 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (ENSG00000165617), score: 0.46 DCLRE1BDNA cross-link repair 1B (ENSG00000118655), score: 0.52 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (ENSG00000100201), score: 0.49 DDX46DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 (ENSG00000145833), score: 0.4 DDX51DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 (ENSG00000185163), score: 0.39 DFNB31deafness, autosomal recessive 31 (ENSG00000095397), score: 0.49 DHRS13dehydrogenase/reductase (SDR family) member 13 (ENSG00000167536), score: 0.59 DHRS3dehydrogenase/reductase (SDR family) member 3 (ENSG00000162496), score: -0.54 DIAPH2diaphanous homolog 2 (Drosophila) (ENSG00000147202), score: -0.45 DICER1dicer 1, ribonuclease type III (ENSG00000100697), score: 0.41 DNAJC10DnaJ (Hsp40) homolog, subfamily C, member 10 (ENSG00000077232), score: -0.55 DNM3dynamin 3 (ENSG00000197959), score: 0.43 DPCDdeleted in primary ciliary dyskinesia homolog (mouse) (ENSG00000166171), score: -0.5 DPM1dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit (ENSG00000000419), score: -0.49 E2F3E2F transcription factor 3 (ENSG00000112242), score: -0.48 EBF1early B-cell factor 1 (ENSG00000164330), score: 0.4 EEDembryonic ectoderm development (ENSG00000074266), score: 0.46 ELF1E74-like factor 1 (ets domain transcription factor) (ENSG00000120690), score: -0.48 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: 0.49 ERBB2IPerbb2 interacting protein (ENSG00000112851), score: -0.44 ERP44endoplasmic reticulum protein 44 (ENSG00000023318), score: -0.47 ESPNLespin-like (ENSG00000144488), score: 0.76 ETV6ets variant 6 (ENSG00000139083), score: -0.7 EXOSC10exosome component 10 (ENSG00000171824), score: 0.47 EXOSC2exosome component 2 (ENSG00000130713), score: 0.4 FAM131Bfamily with sequence similarity 131, member B (ENSG00000159784), score: 0.43 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.69 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.62 FGF9fibroblast growth factor 9 (glia-activating factor) (ENSG00000102678), score: 0.48 FGFR1fibroblast growth factor receptor 1 (ENSG00000077782), score: 0.4 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.5 FKBP5FK506 binding protein 5 (ENSG00000096060), score: -0.6 FLT3fms-related tyrosine kinase 3 (ENSG00000122025), score: 0.59 FNDC3Bfibronectin type III domain containing 3B (ENSG00000075420), score: -0.44 FOXO3forkhead box O3 (ENSG00000118689), score: 0.47 FUT9fucosyltransferase 9 (alpha (1,3) fucosyltransferase) (ENSG00000172461), score: 0.51 FYCO1FYVE and coiled-coil domain containing 1 (ENSG00000163820), score: -0.48 FZD7frizzled homolog 7 (Drosophila) (ENSG00000155760), score: 0.63 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (ENSG00000119514), score: 0.43 GALNT7UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (ENSG00000109586), score: 0.47 GBX2gastrulation brain homeobox 2 (ENSG00000168505), score: 0.44 GCFC1GC-rich sequence DNA-binding factor 1 (ENSG00000159086), score: 0.46 GCM2glial cells missing homolog 2 (Drosophila) (ENSG00000124827), score: 0.8 GDF7growth differentiation factor 7 (ENSG00000143869), score: 0.45 GDNFglial cell derived neurotrophic factor (ENSG00000168621), score: 0.68 GFOD2glucose-fructose oxidoreductase domain containing 2 (ENSG00000141098), score: 0.5 GHITMgrowth hormone inducible transmembrane protein (ENSG00000165678), score: -0.46 GIT2G protein-coupled receptor kinase interacting ArfGAP 2 (ENSG00000139436), score: 0.46 GLCEglucuronic acid epimerase (ENSG00000138604), score: 0.62 GLRA2glycine receptor, alpha 2 (ENSG00000101958), score: 0.42 GPR176G protein-coupled receptor 176 (ENSG00000166073), score: 0.47 GPR83G protein-coupled receptor 83 (ENSG00000123901), score: 0.42 GRIA4glutamate receptor, ionotrophic, AMPA 4 (ENSG00000152578), score: 0.43 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.44 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.45 HAUS3HAUS augmin-like complex, subunit 3 (ENSG00000214367), score: 0.43 HECTD2HECT domain containing 2 (ENSG00000165338), score: 0.4 HEMK1HemK methyltransferase family member 1 (ENSG00000114735), score: -0.53 HINFPhistone H4 transcription factor (ENSG00000172273), score: 0.39 HSCBHscB iron-sulfur cluster co-chaperone homolog (E. coli) (ENSG00000100209), score: -0.45 HTThuntingtin (ENSG00000197386), score: 0.4 IFFO2intermediate filament family orphan 2 (ENSG00000169991), score: 0.48 IL17RAinterleukin 17 receptor A (ENSG00000177663), score: -0.5 IMPG1interphotoreceptor matrix proteoglycan 1 (ENSG00000112706), score: 0.72 JMJD1Cjumonji domain containing 1C (ENSG00000171988), score: 0.64 KCNA1potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) (ENSG00000111262), score: 0.43 KCNC1potassium voltage-gated channel, Shaw-related subfamily, member 1 (ENSG00000129159), score: 0.48 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (ENSG00000171385), score: 0.42 KCNK1potassium channel, subfamily K, member 1 (ENSG00000135750), score: 0.41 KCNK10potassium channel, subfamily K, member 10 (ENSG00000100433), score: 0.61 KCNK9potassium channel, subfamily K, member 9 (ENSG00000169427), score: 0.56 KCNQ2potassium voltage-gated channel, KQT-like subfamily, member 2 (ENSG00000075043), score: 0.39 KCNRGpotassium channel regulator (ENSG00000198553), score: 0.77 KCNT1potassium channel, subfamily T, member 1 (ENSG00000107147), score: 0.48 KCTD2potassium channel tetramerisation domain containing 2 (ENSG00000180901), score: 0.57 KDM3Blysine (K)-specific demethylase 3B (ENSG00000120733), score: 0.4 KIAA0922KIAA0922 (ENSG00000121210), score: -0.54 KIAA1217KIAA1217 (ENSG00000120549), score: -0.57 KIAA1614KIAA1614 (ENSG00000135835), score: 0.4 KIF13Bkinesin family member 13B (ENSG00000197892), score: -0.46 KLF7Kruppel-like factor 7 (ubiquitous) (ENSG00000118263), score: 0.39 KRT222keratin 222 (ENSG00000213424), score: 0.45 LAMC2laminin, gamma 2 (ENSG00000058085), score: 0.54 LBHlimb bud and heart development homolog (mouse) (ENSG00000213626), score: -0.55 LDB2LIM domain binding 2 (ENSG00000169744), score: -0.58 LDLRAP1low density lipoprotein receptor adaptor protein 1 (ENSG00000157978), score: 0.56 LHX4LIM homeobox 4 (ENSG00000121454), score: 0.43 LIMA1LIM domain and actin binding 1 (ENSG00000050405), score: 0.41 LOC100134209similar to IQ motif and Sec7 domain-containing protein 3 (ENSG00000120645), score: 0.4 LRCH1leucine-rich repeats and calponin homology (CH) domain containing 1 (ENSG00000136141), score: 0.64 LRRC3Bleucine rich repeat containing 3B (ENSG00000179796), score: 0.4 LYRM5LYR motif containing 5 (ENSG00000205707), score: -0.55 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.73 MAML1mastermind-like 1 (Drosophila) (ENSG00000161021), score: 0.58 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.6 MAN2B2mannosidase, alpha, class 2B, member 2 (ENSG00000013288), score: -0.53 MARK3MAP/microtubule affinity-regulating kinase 3 (ENSG00000075413), score: 0.48 MCF2LMCF.2 cell line derived transforming sequence-like (ENSG00000126217), score: 0.39 MCTP1multiple C2 domains, transmembrane 1 (ENSG00000175471), score: 0.42 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.79 MED13Lmediator complex subunit 13-like (ENSG00000123066), score: 0.45 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.46 MGAT4Amannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A (ENSG00000071073), score: 0.47 MID1midline 1 (Opitz/BBB syndrome) (ENSG00000101871), score: 0.56 MIPEPmitochondrial intermediate peptidase (ENSG00000027001), score: -0.49 MLLmyeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) (ENSG00000118058), score: 0.52 MRP63mitochondrial ribosomal protein 63 (ENSG00000173141), score: -0.6 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: -0.53 MRPS16mitochondrial ribosomal protein S16 (ENSG00000182180), score: -0.56 MRPS25mitochondrial ribosomal protein S25 (ENSG00000131368), score: -0.49 MTAPmethylthioadenosine phosphorylase (ENSG00000099810), score: -0.54 MTIF2mitochondrial translational initiation factor 2 (ENSG00000085760), score: -0.58 MTSS1Lmetastasis suppressor 1-like (ENSG00000132613), score: 0.39 MUC6mucin 6, oligomeric mucus/gel-forming (ENSG00000184956), score: 0.51 MYLIPmyosin regulatory light chain interacting protein (ENSG00000007944), score: -0.47 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.67 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (ENSG00000103174), score: -0.55 NEIL1nei endonuclease VIII-like 1 (E. coli) (ENSG00000140398), score: 0.44 NFKBIDnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta (ENSG00000167604), score: 0.4 NIPAL3NIPA-like domain containing 3 (ENSG00000001461), score: 0.4 NKX6-3NK6 homeobox 3 (ENSG00000165066), score: 1 NR1H3nuclear receptor subfamily 1, group H, member 3 (ENSG00000025434), score: -0.45 NR2C2nuclear receptor subfamily 2, group C, member 2 (ENSG00000177463), score: 0.4 NRP1neuropilin 1 (ENSG00000099250), score: -0.47 NTF3neurotrophin 3 (ENSG00000185652), score: 0.47 NUDCD2NudC domain containing 2 (ENSG00000170584), score: -0.5 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: -0.44 PAG1phosphoprotein associated with glycosphingolipid microdomains 1 (ENSG00000076641), score: 0.43 PAK7p21 protein (Cdc42/Rac)-activated kinase 7 (ENSG00000101349), score: 0.39 PANK3pantothenate kinase 3 (ENSG00000120137), score: 0.4 PARP12poly (ADP-ribose) polymerase family, member 12 (ENSG00000059378), score: -0.5 PAX6paired box 6 (ENSG00000007372), score: 0.66 PAXIP1PAX interacting (with transcription-activation domain) protein 1 (ENSG00000157212), score: 0.5 PCYOX1prenylcysteine oxidase 1 (ENSG00000116005), score: -0.45 PDE9Aphosphodiesterase 9A (ENSG00000160191), score: 0.39 PDIA3protein disulfide isomerase family A, member 3 (ENSG00000167004), score: -0.45 PDZD7PDZ domain containing 7 (ENSG00000186862), score: 0.4 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: -0.55 PFKFB26-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (ENSG00000123836), score: -0.44 PHF5APHD finger protein 5A (ENSG00000100410), score: -0.52 PHIPpleckstrin homology domain interacting protein (ENSG00000146247), score: 0.58 PIGPphosphatidylinositol glycan anchor biosynthesis, class P (ENSG00000185808), score: -0.55 PIK3R3phosphoinositide-3-kinase, regulatory subunit 3 (gamma) (ENSG00000117461), score: 0.39 PKNOX1PBX/knotted 1 homeobox 1 (ENSG00000160199), score: 0.43 PLCB4phospholipase C, beta 4 (ENSG00000101333), score: 0.5 PLCXD3phosphatidylinositol-specific phospholipase C, X domain containing 3 (ENSG00000182836), score: 0.52 PLD5phospholipase D family, member 5 (ENSG00000180287), score: 0.49 PLEKHF2pleckstrin homology domain containing, family F (with FYVE domain) member 2 (ENSG00000175895), score: -0.53 PLK2polo-like kinase 2 (ENSG00000145632), score: -0.55 POLBpolymerase (DNA directed), beta (ENSG00000070501), score: 0.48 POLG2polymerase (DNA directed), gamma 2, accessory subunit (ENSG00000136480), score: 0.52 POP5processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) (ENSG00000167272), score: -0.59 PPIGpeptidylprolyl isomerase G (cyclophilin G) (ENSG00000138398), score: 0.44 PPIL3peptidylprolyl isomerase (cyclophilin)-like 3 (ENSG00000240344), score: -0.5 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha (ENSG00000066027), score: -0.54 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.41 PRPF38BPRP38 pre-mRNA processing factor 38 (yeast) domain containing B (ENSG00000134186), score: 0.56 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (ENSG00000101161), score: 0.39 PTCH1patched 1 (ENSG00000185920), score: 0.57 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.51 PTK2BPTK2B protein tyrosine kinase 2 beta (ENSG00000120899), score: 0.42 PVRL3poliovirus receptor-related 3 (ENSG00000177707), score: -0.45 PYGO1pygopus homolog 1 (Drosophila) (ENSG00000171016), score: 0.42 QSOX2quiescin Q6 sulfhydryl oxidase 2 (ENSG00000165661), score: 0.58 RAB20RAB20, member RAS oncogene family (ENSG00000139832), score: -0.46 RAB26RAB26, member RAS oncogene family (ENSG00000167964), score: 0.48 RAB9BRAB9B, member RAS oncogene family (ENSG00000123570), score: 0.39 RAPGEF6Rap guanine nucleotide exchange factor (GEF) 6 (ENSG00000158987), score: 0.39 RASGEF1CRasGEF domain family, member 1C (ENSG00000146090), score: 0.41 RBM6RNA binding motif protein 6 (ENSG00000004534), score: 0.44 RBM9RNA binding motif protein 9 (ENSG00000100320), score: 0.4 RFTN1raftlin, lipid raft linker 1 (ENSG00000131378), score: -0.5 RGS11regulator of G-protein signaling 11 (ENSG00000076344), score: 0.44 RHBDD1rhomboid domain containing 1 (ENSG00000144468), score: -0.46 RIN2Ras and Rab interactor 2 (ENSG00000132669), score: -0.49 RIPK2receptor-interacting serine-threonine kinase 2 (ENSG00000104312), score: -0.55 RIT2Ras-like without CAAX 2 (ENSG00000152214), score: 0.55 RLFrearranged L-myc fusion (ENSG00000117000), score: 0.39 RMND5Arequired for meiotic nuclear division 5 homolog A (S. cerevisiae) (ENSG00000153561), score: 0.54 RND3Rho family GTPase 3 (ENSG00000115963), score: -0.46 RNF122ring finger protein 122 (ENSG00000133874), score: 0.42 RNF144Aring finger protein 144A (ENSG00000151692), score: 0.42 RNF152ring finger protein 152 (ENSG00000176641), score: 0.44 ROBLD3roadblock domain containing 3 (ENSG00000116586), score: -0.47 RSBN1round spermatid basic protein 1 (ENSG00000081019), score: 0.41 RUNX1T1runt-related transcription factor 1; translocated to, 1 (cyclin D-related) (ENSG00000079102), score: 0.45 SCGNsecretagogin, EF-hand calcium binding protein (ENSG00000079689), score: 0.46 SCRN1secernin 1 (ENSG00000136193), score: 0.42 SEC31BSEC31 homolog B (S. cerevisiae) (ENSG00000075826), score: 0.46 SEL1L3sel-1 suppressor of lin-12-like 3 (C. elegans) (ENSG00000091490), score: 0.42 SENP7SUMO1/sentrin specific peptidase 7 (ENSG00000138468), score: 0.45 SETBP1SET binding protein 1 (ENSG00000152217), score: 0.45 SETD5SET domain containing 5 (ENSG00000168137), score: 0.49 SF3B3splicing factor 3b, subunit 3, 130kDa (ENSG00000189091), score: 0.55 SFPQsplicing factor proline/glutamine-rich (ENSG00000116560), score: 0.49 SFRS18splicing factor, arginine/serine-rich 18 (ENSG00000132424), score: 0.53 SFXN3sideroflexin 3 (ENSG00000107819), score: 0.4 SHFSrc homology 2 domain containing F (ENSG00000138606), score: 0.46 SIDT1SID1 transmembrane family, member 1 (ENSG00000072858), score: 0.46 SKAP2src kinase associated phosphoprotein 2 (ENSG00000005020), score: -0.47 SKIv-ski sarcoma viral oncogene homolog (avian) (ENSG00000157933), score: 0.41 SKOR1SKI family transcriptional corepressor 1 (ENSG00000188779), score: 0.55 SLC30A1solute carrier family 30 (zinc transporter), member 1 (ENSG00000170385), score: -0.51 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.6 SLITRK4SLIT and NTRK-like family, member 4 (ENSG00000179542), score: 0.4 SNCAIPsynuclein, alpha interacting protein (ENSG00000064692), score: 0.48 SNED1sushi, nidogen and EGF-like domains 1 (ENSG00000162804), score: 0.44 SNX25sorting nexin 25 (ENSG00000109762), score: 0.4 SPHKAPSPHK1 interactor, AKAP domain containing (ENSG00000153820), score: 0.44 SPINK6serine peptidase inhibitor, Kazal type 6 (ENSG00000178172), score: 0.77 SPTBN5spectrin, beta, non-erythrocytic 5 (ENSG00000137877), score: 0.67 SRRM1serine/arginine repetitive matrix 1 (ENSG00000133226), score: 0.4 SRRM4serine/arginine repetitive matrix 4 (ENSG00000139767), score: 0.47 SSH1slingshot homolog 1 (Drosophila) (ENSG00000084112), score: 0.42 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: -0.54 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.59 STARD13StAR-related lipid transfer (START) domain containing 13 (ENSG00000133121), score: -0.44 STX16syntaxin 16 (ENSG00000124222), score: 0.42 SUN1Sad1 and UNC84 domain containing 1 (ENSG00000164828), score: 0.43 SUZ12suppressor of zeste 12 homolog (Drosophila) (ENSG00000178691), score: 0.41 SYCP2Lsynaptonemal complex protein 2-like (ENSG00000153157), score: 0.94 SYT12synaptotagmin XII (ENSG00000173227), score: 0.4 SYT4synaptotagmin IV (ENSG00000132872), score: 0.5 TATDN3TatD DNase domain containing 3 (ENSG00000203705), score: -0.44 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (ENSG00000109436), score: 0.43 TEX264testis expressed 264 (ENSG00000164081), score: -0.58 TFAP2Etranscription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) (ENSG00000116819), score: 0.74 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.71 TLL1tolloid-like 1 (ENSG00000038295), score: 0.59 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: -0.5 TMEM63Ctransmembrane protein 63C (ENSG00000165548), score: 0.39 TMEM74transmembrane protein 74 (ENSG00000164841), score: 0.42 TMTC2transmembrane and tetratricopeptide repeat containing 2 (ENSG00000179104), score: 0.48 TNFRSF1Atumor necrosis factor receptor superfamily, member 1A (ENSG00000067182), score: -0.45 TOP2Btopoisomerase (DNA) II beta 180kDa (ENSG00000077097), score: 0.52 TP73tumor protein p73 (ENSG00000078900), score: 0.69 TRHDEthyrotropin-releasing hormone degrading enzyme (ENSG00000072657), score: 0.42 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.6 TSPAN18tetraspanin 18 (ENSG00000157570), score: 0.42 TTC33tetratricopeptide repeat domain 33 (ENSG00000113638), score: 0.41 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (ENSG00000197355), score: -0.45 UNC5Bunc-5 homolog B (C. elegans) (ENSG00000107731), score: 0.4 UNC80unc-80 homolog (C. elegans) (ENSG00000144406), score: 0.4 UPF0639UPF0639 protein (ENSG00000175985), score: 0.5 VASH1vasohibin 1 (ENSG00000071246), score: 0.42 VAT1Lvesicle amine transport protein 1 homolog (T. californica)-like (ENSG00000171724), score: 0.51 VPS45vacuolar protein sorting 45 homolog (S. cerevisiae) (ENSG00000136631), score: 0.4 VSX1visual system homeobox 1 (ENSG00000100987), score: 0.76 WACWW domain containing adaptor with coiled-coil (ENSG00000095787), score: 0.46 WASF3WAS protein family, member 3 (ENSG00000132970), score: 0.4 WDR8WD repeat domain 8 (ENSG00000116213), score: 0.4 WHSC1L1Wolf-Hirschhorn syndrome candidate 1-like 1 (ENSG00000147548), score: 0.41 WHSC2Wolf-Hirschhorn syndrome candidate 2 (ENSG00000185049), score: 0.42 XRN15'-3' exoribonuclease 1 (ENSG00000114127), score: 0.42 YIPF1Yip1 domain family, member 1 (ENSG00000058799), score: -0.54 YPEL4yippee-like 4 (Drosophila) (ENSG00000166793), score: 0.4 ZBTB34zinc finger and BTB domain containing 34 (ENSG00000177125), score: 0.42 ZDHHC21zinc finger, DHHC-type containing 21 (ENSG00000175893), score: 0.47 ZFHX3zinc finger homeobox 3 (ENSG00000140836), score: -0.45 ZFPM2zinc finger protein, multitype 2 (ENSG00000169946), score: 0.45 ZIC4Zic family member 4 (ENSG00000174963), score: 0.7 ZNF238zinc finger protein 238 (ENSG00000179456), score: 0.5 ZNF292zinc finger protein 292 (ENSG00000188994), score: 0.38 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.62 ZNF536zinc finger protein 536 (ENSG00000198597), score: 0.4 ZNF827zinc finger protein 827 (ENSG00000151612), score: 0.38
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |