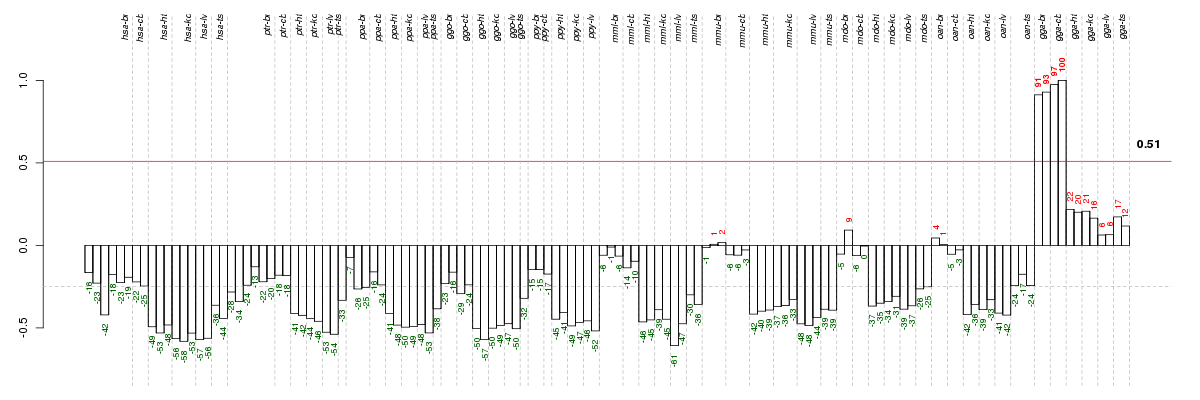

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
ligand-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
A2LD1AIG2-like domain 1 (ENSG00000134864), score: 0.37 AAK1AP2 associated kinase 1 (ENSG00000115977), score: 0.44 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.4 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (ENSG00000156140), score: 0.38 ADARB2adenosine deaminase, RNA-specific, B2 (ENSG00000185736), score: 0.39 ADAT1adenosine deaminase, tRNA-specific 1 (ENSG00000065457), score: 0.42 ADAT2adenosine deaminase, tRNA-specific 2, TAD2 homolog (S. cerevisiae) (ENSG00000189007), score: 0.43 ADNPactivity-dependent neuroprotector homeobox (ENSG00000101126), score: 0.48 ALG14asparagine-linked glycosylation 14 homolog (S. cerevisiae) (ENSG00000172339), score: -0.37 ALX4ALX homeobox 4 (ENSG00000052850), score: 0.73 AMHanti-Mullerian hormone (ENSG00000104899), score: 0.41 AMOTangiomotin (ENSG00000126016), score: 0.5 ANGPT2angiopoietin 2 (ENSG00000091879), score: 0.65 ANKRA2ankyrin repeat, family A (RFXANK-like), 2 (ENSG00000164331), score: 0.44 ANKRD42ankyrin repeat domain 42 (ENSG00000137494), score: -0.37 ANKRD44ankyrin repeat domain 44 (ENSG00000065413), score: 0.58 ANO5anoctamin 5 (ENSG00000171714), score: 0.38 APBA3amyloid beta (A4) precursor protein-binding, family A, member 3 (ENSG00000011132), score: -0.38 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (ENSG00000149182), score: -0.42 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: -0.36 ARFRP1ADP-ribosylation factor related protein 1 (ENSG00000101246), score: -0.39 ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: -0.51 ARPP21cAMP-regulated phosphoprotein, 21kDa (ENSG00000172995), score: 0.42 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.39 ATP13A5ATPase type 13A5 (ENSG00000187527), score: 0.65 AURKAIP1aurora kinase A interacting protein 1 (ENSG00000175756), score: -0.39 AXIN2axin 2 (ENSG00000168646), score: 0.39 B3GALT1UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 (ENSG00000172318), score: 0.39 BACE1beta-site APP-cleaving enzyme 1 (ENSG00000186318), score: 0.4 BAHCC1BAH domain and coiled-coil containing 1 (ENSG00000171282), score: 0.44 BAT2L2HLA-B associated transcript 2-like 2 (ENSG00000117523), score: 0.5 BET3LBET3 like (S. cerevisiae) (ENSG00000173626), score: 0.75 BLKB lymphoid tyrosine kinase (ENSG00000136573), score: 0.69 BMP15bone morphogenetic protein 15 (ENSG00000130385), score: 0.42 BPIL3bactericidal/permeability-increasing protein-like 3 (ENSG00000167104), score: 0.61 BRS3bombesin-like receptor 3 (ENSG00000102239), score: 0.62 BRWD3bromodomain and WD repeat domain containing 3 (ENSG00000165288), score: 0.47 C11orf74chromosome 11 open reading frame 74 (ENSG00000166352), score: -0.38 C13orf39chromosome 13 open reading frame 39 (ENSG00000139780), score: 0.5 C16orf42chromosome 16 open reading frame 42 (ENSG00000007520), score: -0.44 C16orf68chromosome 16 open reading frame 68 (ENSG00000067365), score: -0.37 C17orf75chromosome 17 open reading frame 75 (ENSG00000108666), score: 0.39 C18orf22chromosome 18 open reading frame 22 (ENSG00000101546), score: -0.55 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.39 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: -0.48 C21orf29chromosome 21 open reading frame 29 (ENSG00000175894), score: 0.46 C3orf17chromosome 3 open reading frame 17 (ENSG00000163608), score: -0.5 C5orf24chromosome 5 open reading frame 24 (ENSG00000181904), score: 0.5 C6orf162chromosome 6 open reading frame 162 (ENSG00000111850), score: -0.37 C7orf16chromosome 7 open reading frame 16 (ENSG00000106341), score: 0.44 C9orf91chromosome 9 open reading frame 91 (ENSG00000157693), score: -0.36 CACHD1cache domain containing 1 (ENSG00000158966), score: 0.48 CACNA1Ecalcium channel, voltage-dependent, R type, alpha 1E subunit (ENSG00000198216), score: 0.38 CALCRLcalcitonin receptor-like (ENSG00000064989), score: 0.39 CAPZBcapping protein (actin filament) muscle Z-line, beta (ENSG00000077549), score: -0.34 CARD9caspase recruitment domain family, member 9 (ENSG00000187796), score: 0.44 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: -0.38 CCDC28Bcoiled-coil domain containing 28B (ENSG00000160050), score: -0.38 CCDC45coiled-coil domain containing 45 (ENSG00000141325), score: -0.4 CCDC77coiled-coil domain containing 77 (ENSG00000120647), score: -0.34 CDH19cadherin 19, type 2 (ENSG00000071991), score: 0.46 CDKL5cyclin-dependent kinase-like 5 (ENSG00000008086), score: 0.39 CDR2cerebellar degeneration-related protein 2, 62kDa (ENSG00000140743), score: -0.41 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: -0.42 CHD6chromodomain helicase DNA binding protein 6 (ENSG00000124177), score: 0.45 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.46 CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.49 CINPcyclin-dependent kinase 2 interacting protein (ENSG00000100865), score: -0.44 CLVS2clavesin 2 (ENSG00000146352), score: 0.37 CMAScytidine monophosphate N-acetylneuraminic acid synthetase (ENSG00000111726), score: -0.35 CNGB1cyclic nucleotide gated channel beta 1 (ENSG00000070729), score: 0.56 CNR1cannabinoid receptor 1 (brain) (ENSG00000118432), score: 0.39 CNTN3contactin 3 (plasmacytoma associated) (ENSG00000113805), score: 0.4 CNTN4contactin 4 (ENSG00000144619), score: 0.38 CNTN5contactin 5 (ENSG00000149972), score: 0.41 COBRA1cofactor of BRCA1 (ENSG00000188986), score: -0.46 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.37 CRTAPcartilage associated protein (ENSG00000170275), score: -0.36 CSMD2CUB and Sushi multiple domains 2 (ENSG00000121904), score: 0.51 CSRNP3cysteine-serine-rich nuclear protein 3 (ENSG00000178662), score: 0.39 CTTNBP2NLCTTNBP2 N-terminal like (ENSG00000143079), score: 0.4 CUL4Acullin 4A (ENSG00000139842), score: -0.53 CYTL1cytokine-like 1 (ENSG00000170891), score: 0.39 D2HGDHD-2-hydroxyglutarate dehydrogenase (ENSG00000180902), score: -0.35 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.4 DDX24DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 (ENSG00000089737), score: -0.38 DDX31DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 (ENSG00000125485), score: 0.43 DDX55DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 (ENSG00000111364), score: -0.34 DFFBDNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) (ENSG00000169598), score: 0.46 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.51 DHRS13dehydrogenase/reductase (SDR family) member 13 (ENSG00000167536), score: -0.35 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: -0.54 DMAP1DNA methyltransferase 1 associated protein 1 (ENSG00000178028), score: -0.34 DNAJC9DnaJ (Hsp40) homolog, subfamily C, member 9 (ENSG00000213551), score: -0.35 DNMT3ADNA (cytosine-5-)-methyltransferase 3 alpha (ENSG00000119772), score: 0.48 DOCK11dedicator of cytokinesis 11 (ENSG00000147251), score: 0.61 DSCAMDown syndrome cell adhesion molecule (ENSG00000171587), score: 0.42 EGFL6EGF-like-domain, multiple 6 (ENSG00000198759), score: 0.45 ELP2elongation protein 2 homolog (S. cerevisiae) (ENSG00000134759), score: -0.38 ELP4elongation protein 4 homolog (S. cerevisiae) (ENSG00000109911), score: -0.37 EN1engrailed homeobox 1 (ENSG00000163064), score: 0.53 EPGNepithelial mitogen homolog (mouse) (ENSG00000182585), score: 0.58 EPHA3EPH receptor A3 (ENSG00000044524), score: 0.59 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.42 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (ENSG00000143819), score: -0.36 ERAL1Era G-protein-like 1 (E. coli) (ENSG00000132591), score: -0.4 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (ENSG00000178568), score: 0.49 ERGIC3ERGIC and golgi 3 (ENSG00000125991), score: -0.38 EXOSC7exosome component 7 (ENSG00000075914), score: -0.38 FAM105Afamily with sequence similarity 105, member A (ENSG00000145569), score: 0.39 FAM105Bfamily with sequence similarity 105, member B (ENSG00000154124), score: -0.49 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.38 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: 0.51 FAM166Afamily with sequence similarity 166, member A (ENSG00000188163), score: -0.47 FAM54Bfamily with sequence similarity 54, member B (ENSG00000117640), score: -0.38 FAM65Bfamily with sequence similarity 65, member B (ENSG00000111913), score: 0.42 FANCAFanconi anemia, complementation group A (ENSG00000187741), score: -0.4 FBXL18F-box and leucine-rich repeat protein 18 (ENSG00000155034), score: 0.54 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.41 FIP1L1FIP1 like 1 (S. cerevisiae) (ENSG00000145216), score: -0.53 FNBP4formin binding protein 4 (ENSG00000109920), score: 0.44 FOXL2forkhead box L2 (ENSG00000183770), score: 0.49 GAAglucosidase, alpha; acid (ENSG00000171298), score: -0.39 GABRA4gamma-aminobutyric acid (GABA) A receptor, alpha 4 (ENSG00000109158), score: 0.44 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.53 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (ENSG00000111886), score: 0.77 GAD2glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) (ENSG00000136750), score: 0.4 GALNTL6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 (ENSG00000174473), score: 0.41 GAMTguanidinoacetate N-methyltransferase (ENSG00000130005), score: -0.37 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (ENSG00000171766), score: -0.49 GFRALGDNF family receptor alpha like (ENSG00000187871), score: 0.66 GLRA1glycine receptor, alpha 1 (ENSG00000145888), score: 0.38 GLRA2glycine receptor, alpha 2 (ENSG00000101958), score: 0.44 GLRA4glycine receptor, alpha 4 (ENSG00000188828), score: 0.66 GORASP1golgi reassembly stacking protein 1, 65kDa (ENSG00000114745), score: -0.4 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 0.68 GPRC5CG protein-coupled receptor, family C, group 5, member C (ENSG00000170412), score: -0.35 GPX7glutathione peroxidase 7 (ENSG00000116157), score: -0.4 GRAMD3GRAM domain containing 3 (ENSG00000155324), score: -0.34 GRIK1glutamate receptor, ionotropic, kainate 1 (ENSG00000171189), score: 0.63 GRIN3Aglutamate receptor, ionotropic, N-methyl-D-aspartate 3A (ENSG00000198785), score: 0.55 GRPEL2GrpE-like 2, mitochondrial (E. coli) (ENSG00000164284), score: 0.41 GTPBP5GTP binding protein 5 (putative) (ENSG00000101181), score: -0.35 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: -0.35 HELBhelicase (DNA) B (ENSG00000127311), score: 0.43 HELZhelicase with zinc finger (ENSG00000198265), score: 0.46 HMOX2heme oxygenase (decycling) 2 (ENSG00000103415), score: -0.37 HNRNPA0heterogeneous nuclear ribonucleoprotein A0 (ENSG00000177733), score: -0.52 IL1RAPL2interleukin 1 receptor accessory protein-like 2 (ENSG00000189108), score: 0.51 ILDR2immunoglobulin-like domain containing receptor 2 (ENSG00000143195), score: 0.43 IMPG2interphotoreceptor matrix proteoglycan 2 (ENSG00000081148), score: 0.44 INO80DINO80 complex subunit D (ENSG00000114933), score: 0.4 INPPL1inositol polyphosphate phosphatase-like 1 (ENSG00000165458), score: -0.6 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: 0.4 JARID2jumonji, AT rich interactive domain 2 (ENSG00000008083), score: 0.43 KATNA1katanin p60 (ATPase-containing) subunit A 1 (ENSG00000186625), score: -0.41 KCNA10potassium voltage-gated channel, shaker-related subfamily, member 10 (ENSG00000143105), score: 0.85 KCNA3potassium voltage-gated channel, shaker-related subfamily, member 3 (ENSG00000177272), score: 0.45 KCNH7potassium voltage-gated channel, subfamily H (eag-related), member 7 (ENSG00000184611), score: 0.38 KCNJ5potassium inwardly-rectifying channel, subfamily J, member 5 (ENSG00000120457), score: 0.42 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.45 KCTD16potassium channel tetramerisation domain containing 16 (ENSG00000183775), score: 0.45 KERAkeratocan (ENSG00000139330), score: 0.9 KIAA0368KIAA0368 (ENSG00000136813), score: -0.38 KIAA2022KIAA2022 (ENSG00000050030), score: 0.52 KLC4kinesin light chain 4 (ENSG00000137171), score: 0.42 KLHDC3kelch domain containing 3 (ENSG00000124702), score: -0.46 KLHL11kelch-like 11 (Drosophila) (ENSG00000178502), score: 0.42 KLHL28kelch-like 28 (Drosophila) (ENSG00000179454), score: 0.4 LAMP3lysosomal-associated membrane protein 3 (ENSG00000078081), score: 0.52 LEMD2LEM domain containing 2 (ENSG00000161904), score: -0.37 LIN7Alin-7 homolog A (C. elegans) (ENSG00000111052), score: 0.54 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: -0.4 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.5 LOC100293817similar to hCG1811893 (ENSG00000144460), score: 0.48 LONP1lon peptidase 1, mitochondrial (ENSG00000196365), score: -0.35 LONRF3LON peptidase N-terminal domain and ring finger 3 (ENSG00000175556), score: 0.43 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (ENSG00000126243), score: 0.37 LRP12low density lipoprotein receptor-related protein 12 (ENSG00000147650), score: 0.4 LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.46 LSM4LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) (ENSG00000130520), score: -0.45 LYPLAL1lysophospholipase-like 1 (ENSG00000143353), score: -0.36 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.4 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: -0.44 MAP3K13mitogen-activated protein kinase kinase kinase 13 (ENSG00000073803), score: 0.41 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: -0.4 MATN2matrilin 2 (ENSG00000132561), score: -0.35 MBNL3muscleblind-like 3 (Drosophila) (ENSG00000076770), score: 0.4 MCF2MCF.2 cell line derived transforming sequence (ENSG00000101977), score: 0.59 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.58 MED8mediator complex subunit 8 (ENSG00000159479), score: -0.41 METTL9methyltransferase like 9 (ENSG00000197006), score: -0.41 MFSD11major facilitator superfamily domain containing 11 (ENSG00000092931), score: -0.37 MID2midline 2 (ENSG00000080561), score: 0.42 MOCS2molybdenum cofactor synthesis 2 (ENSG00000164172), score: -0.39 MRPL51mitochondrial ribosomal protein L51 (ENSG00000111639), score: -0.38 MSL1male-specific lethal 1 homolog (Drosophila) (ENSG00000188895), score: 0.48 MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.42 MTERFD3MTERF domain containing 3 (ENSG00000120832), score: -0.46 MTMR1myotubularin related protein 1 (ENSG00000063601), score: 0.38 MUL1mitochondrial E3 ubiquitin ligase 1 (ENSG00000090432), score: -0.44 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: 0.58 MYBBP1AMYB binding protein (P160) 1a (ENSG00000132382), score: -0.37 MYO16myosin XVI (ENSG00000041515), score: 0.51 MYOGmyogenin (myogenic factor 4) (ENSG00000122180), score: 1 NADKNAD kinase (ENSG00000008130), score: -0.34 NARFLnuclear prelamin A recognition factor-like (ENSG00000103245), score: -0.34 NAT10N-acetyltransferase 10 (GCN5-related) (ENSG00000135372), score: -0.48 NCOA1nuclear receptor coactivator 1 (ENSG00000084676), score: 0.39 NDUFS3NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) (ENSG00000213619), score: -0.35 NEK9NIMA (never in mitosis gene a)- related kinase 9 (ENSG00000119638), score: -0.37 NFAT5nuclear factor of activated T-cells 5, tonicity-responsive (ENSG00000102908), score: 0.44 NOL6nucleolar protein family 6 (RNA-associated) (ENSG00000165271), score: -0.34 NOL7nucleolar protein 7, 27kDa (ENSG00000225921), score: -0.37 NPY2Rneuropeptide Y receptor Y2 (ENSG00000185149), score: 0.52 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: -0.35 NTNG1netrin G1 (ENSG00000162631), score: 0.39 NUSAP1nucleolar and spindle associated protein 1 (ENSG00000137804), score: 0.59 OGG18-oxoguanine DNA glycosylase (ENSG00000114026), score: -0.43 OLFML2Aolfactomedin-like 2A (ENSG00000185585), score: 0.39 OLIG3oligodendrocyte transcription factor 3 (ENSG00000177468), score: 0.69 OPCMLopioid binding protein/cell adhesion molecule-like (ENSG00000183715), score: 0.42 ORC5Lorigin recognition complex, subunit 5-like (yeast) (ENSG00000164815), score: -0.36 OTOAotoancorin (ENSG00000155719), score: 0.52 OTOSotospiralin (ENSG00000178602), score: 0.49 P2RY1purinergic receptor P2Y, G-protein coupled, 1 (ENSG00000169860), score: 0.61 PAK7p21 protein (Cdc42/Rac)-activated kinase 7 (ENSG00000101349), score: 0.4 PANK3pantothenate kinase 3 (ENSG00000120137), score: 0.41 PANX3pannexin 3 (ENSG00000154143), score: 0.57 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.57 PDCLphosducin-like (ENSG00000136940), score: 0.56 PDE12phosphodiesterase 12 (ENSG00000174840), score: -0.37 PHF15PHD finger protein 15 (ENSG00000043143), score: 0.44 PHLDA3pleckstrin homology-like domain, family A, member 3 (ENSG00000174307), score: -0.4 PIK3CGphosphoinositide-3-kinase, catalytic, gamma polypeptide (ENSG00000105851), score: 0.5 PITPNC1phosphatidylinositol transfer protein, cytoplasmic 1 (ENSG00000154217), score: 0.39 PKDCCprotein kinase domain containing, cytoplasmic homolog (mouse) (ENSG00000162878), score: -0.34 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (ENSG00000116711), score: 0.58 PLEKHO2pleckstrin homology domain containing, family O member 2 (ENSG00000241839), score: -0.46 POF1Bpremature ovarian failure, 1B (ENSG00000124429), score: 0.42 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (ENSG00000100479), score: 0.5 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (ENSG00000113356), score: 0.43 POU1F1POU class 1 homeobox 1 (ENSG00000064835), score: 0.74 PPP6Cprotein phosphatase 6, catalytic subunit (ENSG00000119414), score: -0.34 PPYR1pancreatic polypeptide receptor 1 (ENSG00000204174), score: 0.5 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.35 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.46 PRKAG3protein kinase, AMP-activated, gamma 3 non-catalytic subunit (ENSG00000115592), score: 0.55 PRKCAprotein kinase C, alpha (ENSG00000154229), score: 0.42 PRKG2protein kinase, cGMP-dependent, type II (ENSG00000138669), score: 0.52 PROM1prominin 1 (ENSG00000007062), score: 0.38 PRSS12protease, serine, 12 (neurotrypsin, motopsin) (ENSG00000164099), score: 0.39 PSEN1presenilin 1 (ENSG00000080815), score: -0.36 PTCD1pentatricopeptide repeat domain 1 (ENSG00000106246), score: -0.37 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: -0.36 QDPRquinoid dihydropteridine reductase (ENSG00000151552), score: -0.42 RAB30RAB30, member RAS oncogene family (ENSG00000137502), score: 0.38 RAB33BRAB33B, member RAS oncogene family (ENSG00000172007), score: 0.37 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.38 RAD54L2RAD54-like 2 (S. cerevisiae) (ENSG00000164080), score: 0.46 RAPGEF6Rap guanine nucleotide exchange factor (GEF) 6 (ENSG00000158987), score: 0.44 RBM26RNA binding motif protein 26 (ENSG00000139746), score: 0.38 RBP1retinol binding protein 1, cellular (ENSG00000114115), score: -0.46 RC3H2ring finger and CCCH-type zinc finger domains 2 (ENSG00000056586), score: 0.49 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.5 RHOrhodopsin (ENSG00000163914), score: 0.57 RNASELribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) (ENSG00000135828), score: 0.38 RPAINRPA interacting protein (ENSG00000129197), score: -0.39 RRHretinal pigment epithelium-derived rhodopsin homolog (ENSG00000180245), score: 0.41 RSBN1Lround spermatid basic protein 1-like (ENSG00000187257), score: 0.38 RSPO3R-spondin 3 homolog (Xenopus laevis) (ENSG00000146374), score: 0.43 SAMD12sterile alpha motif domain containing 12 (ENSG00000177570), score: 0.48 SDF4stromal cell derived factor 4 (ENSG00000078808), score: -0.42 SDK2sidekick homolog 2 (chicken) (ENSG00000069188), score: 0.58 SEMA3Asema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A (ENSG00000075213), score: 0.54 SEMA3Dsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D (ENSG00000153993), score: 0.4 SEMA5Asema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A (ENSG00000112902), score: 0.41 SENP6SUMO1/sentrin specific peptidase 6 (ENSG00000112701), score: 0.42 SEP1515 kDa selenoprotein (ENSG00000183291), score: -0.46 SERTAD4SERTA domain containing 4 (ENSG00000082497), score: 0.39 SESTD1SEC14 and spectrin domains 1 (ENSG00000187231), score: 0.41 SGCZsarcoglycan, zeta (ENSG00000185053), score: 0.53 SIKE1suppressor of IKBKE 1 (ENSG00000052723), score: 0.42 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (ENSG00000204351), score: -0.35 SLBPstem-loop binding protein (ENSG00000163950), score: -0.66 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (ENSG00000117479), score: -0.4 SLC25A37solute carrier family 25, member 37 (ENSG00000147454), score: -0.47 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.43 SLC31A2solute carrier family 31 (copper transporters), member 2 (ENSG00000136867), score: -0.35 SLC5A7solute carrier family 5 (choline transporter), member 7 (ENSG00000115665), score: 0.43 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.44 SLC7A14solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 (ENSG00000013293), score: 0.44 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.44 SNRPA1small nuclear ribonucleoprotein polypeptide A' (ENSG00000131876), score: -0.41 SNTG1syntrophin, gamma 1 (ENSG00000147481), score: 0.39 SNX25sorting nexin 25 (ENSG00000109762), score: 0.46 SNX3sorting nexin 3 (ENSG00000112335), score: -0.54 SPP1secreted phosphoprotein 1 (ENSG00000118785), score: -0.36 SPPL2Bsignal peptide peptidase-like 2B (ENSG00000005206), score: -0.36 SPRED1sprouty-related, EVH1 domain containing 1 (ENSG00000166068), score: 0.47 SQSTM1sequestosome 1 (ENSG00000161011), score: -0.34 SRRM1serine/arginine repetitive matrix 1 (ENSG00000133226), score: 0.51 SSNA1Sjogren syndrome nuclear autoantigen 1 (ENSG00000176101), score: -0.5 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.4 STARD3StAR-related lipid transfer (START) domain containing 3 (ENSG00000131748), score: -0.36 STK11serine/threonine kinase 11 (ENSG00000118046), score: -0.37 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.59 SUCLG1succinate-CoA ligase, alpha subunit (ENSG00000163541), score: -0.35 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: -0.35 SURF1surfeit 1 (ENSG00000148290), score: -0.45 SYNPRsynaptoporin (ENSG00000163630), score: 0.39 SYPL1synaptophysin-like 1 (ENSG00000008282), score: -0.6 TT, brachyury homolog (mouse) (ENSG00000164458), score: 0.74 TAB3TGF-beta activated kinase 1/MAP3K7 binding protein 3 (ENSG00000157625), score: 0.4 TAF5TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa (ENSG00000148835), score: 0.5 TAL2T-cell acute lymphocytic leukemia 2 (ENSG00000186051), score: 0.44 TAOK1TAO kinase 1 (ENSG00000160551), score: 0.44 TBC1D30TBC1 domain family, member 30 (ENSG00000111490), score: 0.39 TBX18T-box 18 (ENSG00000112837), score: 0.39 TCF12transcription factor 12 (ENSG00000140262), score: 0.41 THSD7Athrombospondin, type I, domain containing 7A (ENSG00000005108), score: 0.46 TIMM44translocase of inner mitochondrial membrane 44 homolog (yeast) (ENSG00000104980), score: -0.35 TIRAPtoll-interleukin 1 receptor (TIR) domain containing adaptor protein (ENSG00000150455), score: 0.38 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: -0.34 TLL1tolloid-like 1 (ENSG00000038295), score: 0.57 TM4SF1transmembrane 4 L six family member 1 (ENSG00000169908), score: -0.41 TMC1transmembrane channel-like 1 (ENSG00000165091), score: 0.41 TMEM22transmembrane protein 22 (ENSG00000168917), score: 0.48 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.4 TMEM43transmembrane protein 43 (ENSG00000170876), score: -0.39 TMTC3transmembrane and tetratricopeptide repeat containing 3 (ENSG00000139324), score: 0.47 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: -0.53 TNIKTRAF2 and NCK interacting kinase (ENSG00000154310), score: 0.47 TOR2Atorsin family 2, member A (ENSG00000160404), score: -0.43 TRABDTraB domain containing (ENSG00000170638), score: -0.47 TRPC1transient receptor potential cation channel, subfamily C, member 1 (ENSG00000144935), score: 0.42 TRPC3transient receptor potential cation channel, subfamily C, member 3 (ENSG00000138741), score: 0.4 TRPM2transient receptor potential cation channel, subfamily M, member 2 (ENSG00000142185), score: 0.41 TSHZ2teashirt zinc finger homeobox 2 (ENSG00000182463), score: 0.38 TSTA3tissue specific transplantation antigen P35B (ENSG00000104522), score: -0.37 TXNRD1thioredoxin reductase 1 (ENSG00000198431), score: -0.38 UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.54 UBE2G2ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) (ENSG00000184787), score: -0.36 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: -0.42 USP2ubiquitin specific peptidase 2 (ENSG00000036672), score: -0.37 USP45ubiquitin specific peptidase 45 (ENSG00000123552), score: 0.41 VTI1Avesicle transport through interaction with t-SNAREs homolog 1A (yeast) (ENSG00000151532), score: 0.45 VWA5B1von Willebrand factor A domain containing 5B1 (ENSG00000158816), score: 0.47 WDFY3WD repeat and FYVE domain containing 3 (ENSG00000163625), score: 0.38 WDR34WD repeat domain 34 (ENSG00000119333), score: -0.38 WDR77WD repeat domain 77 (ENSG00000116455), score: -0.35 WFDC1WAP four-disulfide core domain 1 (ENSG00000103175), score: -0.34 WHSC1L1Wolf-Hirschhorn syndrome candidate 1-like 1 (ENSG00000147548), score: 0.43 WNT16wingless-type MMTV integration site family, member 16 (ENSG00000002745), score: 0.39 WNT4wingless-type MMTV integration site family, member 4 (ENSG00000162552), score: 0.45 WNT7Awingless-type MMTV integration site family, member 7A (ENSG00000154764), score: 0.37 WRNIP1Werner helicase interacting protein 1 (ENSG00000124535), score: -0.44 ZBTB33zinc finger and BTB domain containing 33 (ENSG00000177485), score: 0.4 ZBTB37zinc finger and BTB domain containing 37 (ENSG00000185278), score: 0.44 ZBTB40zinc finger and BTB domain containing 40 (ENSG00000184677), score: -0.42 ZBTB6zinc finger and BTB domain containing 6 (ENSG00000186130), score: 0.43 ZBTB8Bzinc finger and BTB domain containing 8B (ENSG00000215897), score: 0.5 ZC3H12Bzinc finger CCCH-type containing 12B (ENSG00000102053), score: 0.49 ZCCHC4zinc finger, CCHC domain containing 4 (ENSG00000168228), score: 0.46 ZFHX4zinc finger homeobox 4 (ENSG00000091656), score: 0.43 ZFYVE19zinc finger, FYVE domain containing 19 (ENSG00000166140), score: 0.43 ZFYVE21zinc finger, FYVE domain containing 21 (ENSG00000100711), score: -0.35 ZMAT4zinc finger, matrin type 4 (ENSG00000165061), score: 0.4 ZNF828zinc finger protein 828 (ENSG00000198824), score: 0.46 ZP1zona pellucida glycoprotein 1 (sperm receptor) (ENSG00000149506), score: 0.38
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_br_m_ca1 | gga | br | m | _ |
| gga_br_f_ca1 | gga | br | f | _ |
| gga_cb_m_ca1 | gga | cb | m | _ |
| gga_cb_f_ca1 | gga | cb | f | _ |