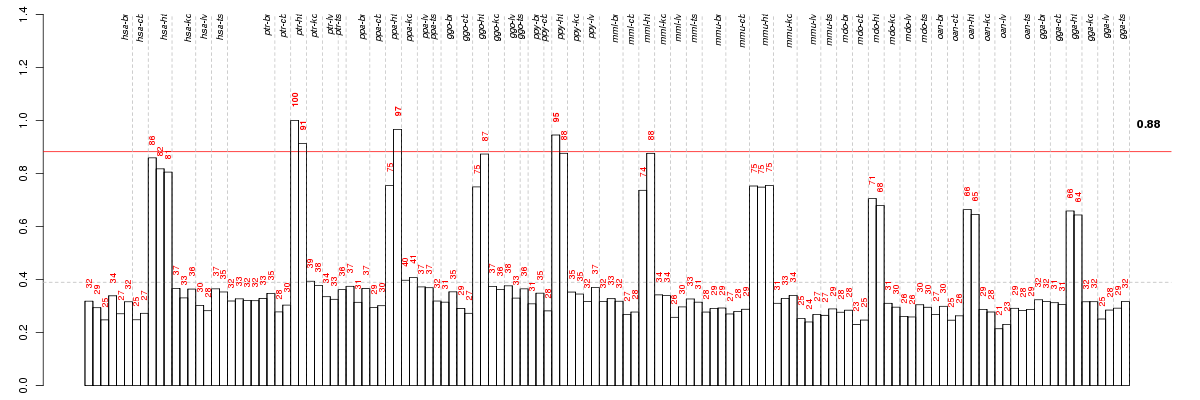

Under-expression is coded with green,
over-expression with red color.

heart morphogenesis
The developmental process by which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
muscle system process
A organ system process carried out at the level of a muscle. Muscle tissue is composed of contractile cells or fibers.
cardiac chamber development
The progression of a cardiac chamber over time, from its formation to the mature structure. A cardiac chamber is an enclosed cavity within the heart.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
cardiac ventricle morphogenesis
The process by which the cardiac ventricle is generated and organized. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
cardiac ventricle development
The process whose specific outcome is the progression of a cardiac ventricle over time, from its formation to the mature structure. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
muscle contraction
A process whereby force is generated within muscle tissue, resulting in a change in muscle geometry. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
heart development
The process whose specific outcome is the progression of the heart over time, from its formation to the mature structure. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
muscle organ development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
actin filament-based movement
Movement of organelles or other particles along actin filaments, or sliding of actin filaments past each other, mediated by motor proteins.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
striated muscle tissue development
The process whose specific outcome is the progression of a striated muscle over time, from its formation to the mature structure. Striated muscle contain fibers that are divided by transverse bands into striations, and cardiac and skeletal muscle are types of striated muscle. Skeletal muscle myoblasts fuse to form myotubes and eventually multinucleated muscle fibers. The fusion of cardiac cells is very rare and can only form binucleate cells.
actin filament-based process
Any cellular process that depends upon or alters the actin cytoskeleton, that part of the cytoskeleton comprising actin filaments and their associated proteins.
muscle filament sliding
The sliding of actin thin filaments and myosin thick filaments past each other in muscle contraction. This involves a process of interaction of myosin located on a thick filament with actin located on a thin filament. During this process ATP is split and forces are generated.
cytoskeleton-dependent intracellular transport
The directed movement of substances along cytoskeletal elements such as microfilaments or microtubules within a cell.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
actin-myosin filament sliding
The sliding movement of actin thin filaments and myosin thick filaments past each other.
intracellular transport
The directed movement of substances within a cell.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cardiac muscle tissue development
The process whose specific outcome is the progression of cardiac muscle over time, from its formation to the mature structure.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
muscle tissue development
The progression of muscle tissue over time, from its initial formation to its mature state. Muscle tissue is a contractile tissue made up of actin and myosin fibers.
muscle structure development
The progression of a muscle structure over time, from its formation to its mature state. Muscle structures are contractile cells, tissues or organs that are found in multicellular organisms.
actin-mediated cell contraction
The actin filament-based process by which cytoplasmic actin filaments slide past one another resulting in contraction of all or part of the cell body.
all
NA
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
intracellular transport
The directed movement of substances within a cell.
actin-myosin filament sliding
The sliding movement of actin thin filaments and myosin thick filaments past each other.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
muscle organ development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
muscle filament sliding
The sliding of actin thin filaments and myosin thick filaments past each other in muscle contraction. This involves a process of interaction of myosin located on a thick filament with actin located on a thin filament. During this process ATP is split and forces are generated.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
cardiac ventricle morphogenesis
The process by which the cardiac ventricle is generated and organized. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
heart morphogenesis
The developmental process by which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
cardiac chamber development
The progression of a cardiac chamber over time, from its formation to the mature structure. A cardiac chamber is an enclosed cavity within the heart.
muscle tissue development
The progression of muscle tissue over time, from its initial formation to its mature state. Muscle tissue is a contractile tissue made up of actin and myosin fibers.
actin filament-based movement
Movement of organelles or other particles along actin filaments, or sliding of actin filaments past each other, mediated by motor proteins.
cardiac muscle tissue development
The process whose specific outcome is the progression of cardiac muscle over time, from its formation to the mature structure.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
striated muscle thin filament
Filaments formed of actin and associated proteins; attached to Z discs at either end of sarcomeres in myofibrils.
adherens junction
A cell junction at which anchoring proteins (cadherins or integrins) extend through the plasma membrane and are attached to actin filaments.
actin cytoskeleton
The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes.
myofibril
The contractile element of skeletal and cardiac muscle; a long, highly organized bundle of actin, myosin, and other proteins that contracts by a sliding filament mechanism.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.
Z disc
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached.
cell junction
A plasma membrane part that forms a specialized region of connection between two cells or between a cell and the extracellular matrix. At a cell junction, anchoring proteins extend through the plasma membrane to link cytoskeletal proteins in one cell to cytoskeletal proteins in neighboring cells or to proteins in the extracellular matrix.
organelle membrane
The lipid bilayer surrounding an organelle.
pseudopodium
A temporary protrusion or retractile process of a cell, associated with flowing movements of the protoplasm, and serving for locomotion and feeding.
I band
A region of a sarcomere that appears as a light band on each side of the Z disc, comprising a region of the sarcomere where thin (actin) filaments are not overlapped by thick (myosin) filaments; contains actin, troponin, and tropomyosin; each sarcomere includes half of an I band at each end.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
envelope
A multilayered structure surrounding all or part of a cell; encompasses one or more lipid bilayers, and may include a cell wall layer; also includes the space between layers.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
anchoring junction
A cell junction that mechanically attaches a cell (and its cytoskeleton) to neighboring cells or to the extracellular matrix.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
striated muscle thin filament
Filaments formed of actin and associated proteins; attached to Z discs at either end of sarcomeres in myofibrils.
striated muscle thin filament
Filaments formed of actin and associated proteins; attached to Z discs at either end of sarcomeres in myofibrils.
I band
A region of a sarcomere that appears as a light band on each side of the Z disc, comprising a region of the sarcomere where thin (actin) filaments are not overlapped by thick (myosin) filaments; contains actin, troponin, and tropomyosin; each sarcomere includes half of an I band at each end.
Z disc
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.
striated muscle thin filament
Filaments formed of actin and associated proteins; attached to Z discs at either end of sarcomeres in myofibrils.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
structural molecule activity
The action of a molecule that contributes to the structural integrity of a complex or assembly within or outside a cell.
structural constituent of muscle
The action of a molecule that contributes to the structural integrity of a muscle fiber.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04260 | 3.039e-05 | 1.674 | 11 | 25 | Cardiac muscle contraction |
| 05410 | 1.124e-04 | 2.276 | 12 | 34 | Hypertrophic cardiomyopathy (HCM) |
| 05414 | 1.124e-04 | 2.276 | 12 | 34 | Dilated cardiomyopathy |
| 05412 | 2.852e-02 | 2.075 | 8 | 31 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) |
ABHD6abhydrolase domain containing 6 (ENSG00000163686), score: -0.46 ABLIM1actin binding LIM protein 1 (ENSG00000099204), score: 0.48 ABRAactin-binding Rho activating protein (ENSG00000174429), score: 0.48 ACTN2actinin, alpha 2 (ENSG00000077522), score: 0.55 ADALadenosine deaminase-like (ENSG00000168803), score: 0.49 ADAMTSL5ADAMTS-like 5 (ENSG00000185761), score: 0.46 ADCY5adenylate cyclase 5 (ENSG00000173175), score: 0.47 ADNPactivity-dependent neuroprotector homeobox (ENSG00000101126), score: -0.44 ADSSL1adenylosuccinate synthase like 1 (ENSG00000185100), score: 0.45 AFAP1L1actin filament associated protein 1-like 1 (ENSG00000157510), score: 0.48 AFTPHaftiphilin (ENSG00000119844), score: -0.46 AIMP1aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 (ENSG00000164022), score: 0.43 AKAP13A kinase (PRKA) anchor protein 13 (ENSG00000170776), score: 0.54 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: -0.45 AMDHD2amidohydrolase domain containing 2 (ENSG00000162066), score: -0.48 ANAPC16anaphase promoting complex subunit 16 (ENSG00000166295), score: 0.43 ANGPTL7angiopoietin-like 7 (ENSG00000171819), score: 0.45 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (ENSG00000148677), score: 0.76 ANKRD2ankyrin repeat domain 2 (stretch responsive muscle) (ENSG00000165887), score: 0.43 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: 0.41 ANO5anoctamin 5 (ENSG00000171714), score: 0.59 ANXA1annexin A1 (ENSG00000135046), score: 0.43 AP1B1adaptor-related protein complex 1, beta 1 subunit (ENSG00000100280), score: -0.44 APOBEC2apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 (ENSG00000124701), score: 0.62 AQP11aquaporin 11 (ENSG00000178301), score: -0.48 ARHGAP10Rho GTPase activating protein 10 (ENSG00000071205), score: 0.46 ARHGEF6Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 (ENSG00000129675), score: 0.51 ARL6IP1ADP-ribosylation factor-like 6 interacting protein 1 (ENSG00000170540), score: -0.67 ARL6IP5ADP-ribosylation-like factor 6 interacting protein 5 (ENSG00000144746), score: 0.48 ASAH1N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ENSG00000104763), score: 0.48 ASB11ankyrin repeat and SOCS box-containing 11 (ENSG00000165192), score: 0.68 ASB14ankyrin repeat and SOCS box-containing 14 (ENSG00000239388), score: 0.44 ASB15ankyrin repeat and SOCS box-containing 15 (ENSG00000146809), score: 0.58 ASB2ankyrin repeat and SOCS box-containing 2 (ENSG00000100628), score: 0.51 ASPNasporin (ENSG00000106819), score: 0.46 ATG13ATG13 autophagy related 13 homolog (S. cerevisiae) (ENSG00000175224), score: -0.45 ATP13A1ATPase type 13A1 (ENSG00000105726), score: -0.59 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (ENSG00000174437), score: 0.44 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: 0.48 ATPIF1ATPase inhibitory factor 1 (ENSG00000130770), score: 0.45 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: 0.5 B4GALNT3beta-1,4-N-acetyl-galactosaminyl transferase 3 (ENSG00000139044), score: 0.42 BCL2L13BCL2-like 13 (apoptosis facilitator) (ENSG00000099968), score: 0.5 BMPR2bone morphogenetic protein receptor, type II (serine/threonine kinase) (ENSG00000204217), score: 0.43 BRI3BPBRI3 binding protein (ENSG00000184992), score: -0.7 C15orf41chromosome 15 open reading frame 41 (ENSG00000186073), score: 0.57 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: 0.49 C19orf28chromosome 19 open reading frame 28 (ENSG00000161091), score: -0.47 C1orf151chromosome 1 open reading frame 151 (ENSG00000173436), score: 0.6 C1QTNF1C1q and tumor necrosis factor related protein 1 (ENSG00000173918), score: 0.44 C4orf32chromosome 4 open reading frame 32 (ENSG00000174749), score: 0.43 C6orf142chromosome 6 open reading frame 142 (ENSG00000146147), score: 0.59 C6orf192chromosome 6 open reading frame 192 (ENSG00000146409), score: -0.49 C7orf27chromosome 7 open reading frame 27 (ENSG00000106009), score: -0.44 C9orf16chromosome 9 open reading frame 16 (ENSG00000171159), score: -0.46 CABLES1Cdk5 and Abl enzyme substrate 1 (ENSG00000134508), score: -0.5 CACNA1Ccalcium channel, voltage-dependent, L type, alpha 1C subunit (ENSG00000151067), score: 0.41 CAMK2Dcalcium/calmodulin-dependent protein kinase II delta (ENSG00000145349), score: 0.43 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (ENSG00000198898), score: 0.62 CASTcalpastatin (ENSG00000153113), score: 0.54 CBLBCas-Br-M (murine) ecotropic retroviral transforming sequence b (ENSG00000114423), score: 0.6 CCDC76coiled-coil domain containing 76 (ENSG00000122435), score: 0.44 CCDC80coiled-coil domain containing 80 (ENSG00000091986), score: 0.48 CCND2cyclin D2 (ENSG00000118971), score: 0.5 CD274CD274 molecule (ENSG00000120217), score: 0.67 CD34CD34 molecule (ENSG00000174059), score: 0.43 CDC37L1cell division cycle 37 homolog (S. cerevisiae)-like 1 (ENSG00000106993), score: 0.43 CDC42EP3CDC42 effector protein (Rho GTPase binding) 3 (ENSG00000163171), score: 0.46 CDH13cadherin 13, H-cadherin (heart) (ENSG00000140945), score: 0.47 CDH19cadherin 19, type 2 (ENSG00000071991), score: 0.43 CDH2cadherin 2, type 1, N-cadherin (neuronal) (ENSG00000170558), score: 0.54 CDK13cyclin-dependent kinase 13 (ENSG00000065883), score: -0.51 CDS1CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 (ENSG00000163624), score: -0.44 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (ENSG00000115816), score: 0.43 CFLARCASP8 and FADD-like apoptosis regulator (ENSG00000003402), score: 0.53 CLIC5chloride intracellular channel 5 (ENSG00000112782), score: 0.59 CMYA5cardiomyopathy associated 5 (ENSG00000164309), score: 0.67 CNIH4cornichon homolog 4 (Drosophila) (ENSG00000143771), score: 0.45 CNNM2cyclin M2 (ENSG00000148842), score: -0.53 COL15A1collagen, type XV, alpha 1 (ENSG00000204291), score: 0.52 COL6A3collagen, type VI, alpha 3 (ENSG00000163359), score: 0.47 COMMD10COMM domain containing 10 (ENSG00000145781), score: 0.44 COX4I1cytochrome c oxidase subunit IV isoform 1 (ENSG00000131143), score: 0.43 CPEB4cytoplasmic polyadenylation element binding protein 4 (ENSG00000113742), score: 0.47 CRYABcrystallin, alpha B (ENSG00000109846), score: 0.42 CSRP3cysteine and glycine-rich protein 3 (cardiac LIM protein) (ENSG00000129170), score: 0.68 CTNNAL1catenin (cadherin-associated protein), alpha-like 1 (ENSG00000119326), score: 0.43 CUL4Acullin 4A (ENSG00000139842), score: 0.44 CYBASC3cytochrome b, ascorbate dependent 3 (ENSG00000162144), score: -0.5 CYHR1cysteine/histidine-rich 1 (ENSG00000187954), score: -0.49 DCP1BDCP1 decapping enzyme homolog B (S. cerevisiae) (ENSG00000151065), score: -0.48 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (ENSG00000150401), score: 0.47 DDA1DET1 and DDB1 associated 1 (ENSG00000130311), score: 0.44 DDX1DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (ENSG00000079785), score: 0.62 DDX46DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 (ENSG00000145833), score: 0.47 DECR12,4-dienoyl CoA reductase 1, mitochondrial (ENSG00000104325), score: 0.43 DLDdihydrolipoamide dehydrogenase (ENSG00000091140), score: 0.45 DNAJC12DnaJ (Hsp40) homolog, subfamily C, member 12 (ENSG00000108176), score: -0.5 DOLPP1dolichyl pyrophosphate phosphatase 1 (ENSG00000167130), score: -0.56 DPTdermatopontin (ENSG00000143196), score: 0.48 DPY19L3dpy-19-like 3 (C. elegans) (ENSG00000178904), score: -0.44 DSPdesmoplakin (ENSG00000096696), score: 0.47 DTWD2DTW domain containing 2 (ENSG00000169570), score: 0.46 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (ENSG00000105865), score: 0.51 DUSP27dual specificity phosphatase 27 (putative) (ENSG00000198842), score: 0.67 DYRK1Adual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (ENSG00000157540), score: 0.47 EDEM2ER degradation enhancer, mannosidase alpha-like 2 (ENSG00000088298), score: -0.48 EDNRAendothelin receptor type A (ENSG00000151617), score: 0.45 EEF2Keukaryotic elongation factor-2 kinase (ENSG00000103319), score: 0.59 EIF3Aeukaryotic translation initiation factor 3, subunit A (ENSG00000107581), score: 0.52 EIF5Beukaryotic translation initiation factor 5B (ENSG00000158417), score: 0.5 EMILIN2elastin microfibril interfacer 2 (ENSG00000132205), score: 0.52 EMP1epithelial membrane protein 1 (ENSG00000134531), score: 0.47 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (ENSG00000138185), score: 0.61 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: 0.46 ERGv-ets erythroblastosis virus E26 oncogene homolog (avian) (ENSG00000157554), score: 0.44 ESYT2extended synaptotagmin-like protein 2 (ENSG00000117868), score: 0.47 ETV5ets variant 5 (ENSG00000244405), score: -0.49 FAAHfatty acid amide hydrolase (ENSG00000117480), score: -0.46 FAM116Bfamily with sequence similarity 116, member B (ENSG00000205593), score: -0.49 FAM129Afamily with sequence similarity 129, member A (ENSG00000135842), score: 0.74 FAM198Bfamily with sequence similarity 198, member B (ENSG00000164125), score: 0.48 FAM36Afamily with sequence similarity 36, member A (ENSG00000203667), score: 0.46 FAM53Afamily with sequence similarity 53, member A (ENSG00000174137), score: -0.44 FASNfatty acid synthase (ENSG00000169710), score: -0.5 FASTKD2FAST kinase domains 2 (ENSG00000118246), score: 0.58 FBLIM1filamin binding LIM protein 1 (ENSG00000162458), score: 0.46 FBN1fibrillin 1 (ENSG00000166147), score: 0.42 FBXL7F-box and leucine-rich repeat protein 7 (ENSG00000183580), score: 0.51 FBXO30F-box protein 30 (ENSG00000118496), score: 0.44 FBXO32F-box protein 32 (ENSG00000156804), score: 0.6 FBXO40F-box protein 40 (ENSG00000163833), score: 0.71 FBXO9F-box protein 9 (ENSG00000112146), score: -0.46 FGF12fibroblast growth factor 12 (ENSG00000114279), score: 0.46 FGF18fibroblast growth factor 18 (ENSG00000156427), score: 0.74 FGFR2fibroblast growth factor receptor 2 (ENSG00000066468), score: -0.52 FGFR3fibroblast growth factor receptor 3 (ENSG00000068078), score: -0.49 FHL2four and a half LIM domains 2 (ENSG00000115641), score: 0.48 FHOD3formin homology 2 domain containing 3 (ENSG00000134775), score: 0.57 FICDFIC domain containing (ENSG00000198855), score: -0.58 FILIP1filamin A interacting protein 1 (ENSG00000118407), score: 0.62 FITM2fat storage-inducing transmembrane protein 2 (ENSG00000197296), score: 0.56 FLCNfolliculin (ENSG00000154803), score: -0.47 FSD2fibronectin type III and SPRY domain containing 2 (ENSG00000186628), score: 0.62 FXR1fragile X mental retardation, autosomal homolog 1 (ENSG00000114416), score: 0.5 FYCO1FYVE and coiled-coil domain containing 1 (ENSG00000163820), score: 0.47 FYTTD1forty-two-three domain containing 1 (ENSG00000122068), score: 0.51 GAB3GRB2-associated binding protein 3 (ENSG00000160219), score: 0.45 GANgigaxonin (ENSG00000127688), score: 0.44 GATA4GATA binding protein 4 (ENSG00000136574), score: 0.48 GCOM1GRINL1A complex locus (ENSG00000137878), score: 0.76 GEMGTP binding protein overexpressed in skeletal muscle (ENSG00000164949), score: 0.52 GFM2G elongation factor, mitochondrial 2 (ENSG00000164347), score: 0.49 GGHgamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (ENSG00000137563), score: -0.67 GHITMgrowth hormone inducible transmembrane protein (ENSG00000165678), score: 0.53 GJA3gap junction protein, alpha 3, 46kDa (ENSG00000121743), score: 0.46 GJB1gap junction protein, beta 1, 32kDa (ENSG00000169562), score: -0.44 GLT8D2glycosyltransferase 8 domain containing 2 (ENSG00000120820), score: 0.43 GOLGA4golgin A4 (ENSG00000144674), score: 0.44 GPNMBglycoprotein (transmembrane) nmb (ENSG00000136235), score: 0.43 GRIP2glutamate receptor interacting protein 2 (ENSG00000144596), score: 0.55 GRK5G protein-coupled receptor kinase 5 (ENSG00000198873), score: 0.48 GUCY1A2guanylate cyclase 1, soluble, alpha 2 (ENSG00000152402), score: 0.56 GXYLT2glucoside xylosyltransferase 2 (ENSG00000172986), score: 0.41 GYG1glycogenin 1 (ENSG00000163754), score: 0.42 HAT1histone acetyltransferase 1 (ENSG00000128708), score: 0.5 HEY2hairy/enhancer-of-split related with YRPW motif 2 (ENSG00000135547), score: 0.54 HGShepatocyte growth factor-regulated tyrosine kinase substrate (ENSG00000185359), score: -0.59 HHATLhedgehog acyltransferase-like (ENSG00000010282), score: 0.45 HIF1ANhypoxia inducible factor 1, alpha subunit inhibitor (ENSG00000166135), score: 0.48 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.44 HMCN1hemicentin 1 (ENSG00000143341), score: 0.5 HMGCR3-hydroxy-3-methylglutaryl-CoA reductase (ENSG00000113161), score: -0.47 IARS2isoleucyl-tRNA synthetase 2, mitochondrial (ENSG00000067704), score: 0.46 IL6interleukin 6 (interferon, beta 2) (ENSG00000136244), score: 0.5 IP6K1inositol hexakisphosphate kinase 1 (ENSG00000176095), score: -0.56 ITGB1BP1integrin beta 1 binding protein 1 (ENSG00000119185), score: 0.45 ITPRIPinositol 1,4,5-triphosphate receptor interacting protein (ENSG00000148841), score: 0.42 KBTBD10kelch repeat and BTB (POZ) domain containing 10 (ENSG00000239474), score: 0.72 KBTBD5kelch repeat and BTB (POZ) domain containing 5 (ENSG00000157119), score: 0.72 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (ENSG00000121361), score: 0.5 KIAA0232KIAA0232 (ENSG00000170871), score: 0.57 KIAA1598KIAA1598 (ENSG00000187164), score: -0.47 KIF13Akinesin family member 13A (ENSG00000137177), score: 0.64 KIF5Bkinesin family member 5B (ENSG00000170759), score: 0.51 KLHDC8Bkelch domain containing 8B (ENSG00000185909), score: 0.5 KLHL24kelch-like 24 (Drosophila) (ENSG00000114796), score: 0.61 KLHL25kelch-like 25 (Drosophila) (ENSG00000183655), score: -0.58 KLHL31kelch-like 31 (Drosophila) (ENSG00000124743), score: 0.69 KLHL38kelch-like 38 (Drosophila) (ENSG00000175946), score: 0.78 KLHL7kelch-like 7 (Drosophila) (ENSG00000122550), score: 0.65 LAMA4laminin, alpha 4 (ENSG00000112769), score: 0.56 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: 0.42 LBHlimb bud and heart development homolog (mouse) (ENSG00000213626), score: 0.46 LDB3LIM domain binding 3 (ENSG00000122367), score: 0.63 LIMS2LIM and senescent cell antigen-like domains 2 (ENSG00000072163), score: 0.44 LMF2lipase maturation factor 2 (ENSG00000100258), score: -0.44 LMOD2leiomodin 2 (cardiac) (ENSG00000170807), score: 0.69 LOC100289938similar to cytoplasmic tRNA 2-thiolation protein 2 (ENSG00000174177), score: -0.47 LOC100291405similar to protein tyrosine phosphatase-like A domain containing 1 (ENSG00000074696), score: -0.48 LOC100291671similar to SH3-binding domain and glutamic acid-rich protein (ENSG00000185437), score: 0.56 LPAR3lysophosphatidic acid receptor 3 (ENSG00000171517), score: 0.61 LPIN2lipin 2 (ENSG00000101577), score: -0.44 LRRC10leucine rich repeat containing 10 (ENSG00000198812), score: 0.56 LRRC14Bleucine rich repeat containing 14B (ENSG00000185028), score: 0.66 LRRC2leucine rich repeat containing 2 (ENSG00000163827), score: 0.63 LRRC20leucine rich repeat containing 20 (ENSG00000172731), score: 0.42 LRRC39leucine rich repeat containing 39 (ENSG00000122477), score: 0.72 MAMDC2MAM domain containing 2 (ENSG00000165072), score: 0.42 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (ENSG00000162889), score: 0.58 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (ENSG00000114738), score: 0.7 MASP1mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) (ENSG00000127241), score: 0.46 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (ENSG00000082212), score: 0.49 MEOX2mesenchyme homeobox 2 (ENSG00000106511), score: 0.67 METRNLmeteorin, glial cell differentiation regulator-like (ENSG00000176845), score: 0.48 METTL11Bmethyltransferase like 11B (ENSG00000203740), score: 1 MFN2mitofusin 2 (ENSG00000116688), score: 0.64 MFSD2Amajor facilitator superfamily domain containing 2A (ENSG00000168389), score: -0.47 MKKSMcKusick-Kaufman syndrome (ENSG00000125863), score: 0.43 MMRN2multimerin 2 (ENSG00000173269), score: 0.44 MPDZmultiple PDZ domain protein (ENSG00000107186), score: 0.44 MRPL15mitochondrial ribosomal protein L15 (ENSG00000137547), score: 0.48 MRPL22mitochondrial ribosomal protein L22 (ENSG00000082515), score: 0.5 MRPL51mitochondrial ribosomal protein L51 (ENSG00000111639), score: 0.42 MRPS23mitochondrial ribosomal protein S23 (ENSG00000181610), score: 0.44 MRPS9mitochondrial ribosomal protein S9 (ENSG00000135972), score: 0.57 MTERFD3MTERF domain containing 3 (ENSG00000120832), score: 0.42 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: 0.47 MTUS1microtubule associated tumor suppressor 1 (ENSG00000129422), score: 0.41 MYBPC3myosin binding protein C, cardiac (ENSG00000134571), score: 0.61 MYCT1myc target 1 (ENSG00000120279), score: 0.46 MYL3myosin, light chain 3, alkali; ventricular, skeletal, slow (ENSG00000160808), score: 0.62 MYLK3myosin light chain kinase 3 (ENSG00000140795), score: 0.56 MYLK4myosin light chain kinase family, member 4 (ENSG00000145949), score: 0.7 MYOCmyocilin, trabecular meshwork inducible glucocorticoid response (ENSG00000034971), score: 0.63 MYOCDmyocardin (ENSG00000141052), score: 0.6 MYOM2myomesin (M-protein) 2, 165kDa (ENSG00000036448), score: 0.55 MYOM3myomesin family, member 3 (ENSG00000142661), score: 0.75 MYOZ1myozenin 1 (ENSG00000177791), score: 0.57 MYOZ2myozenin 2 (ENSG00000172399), score: 0.71 MYPNmyopalladin (ENSG00000138347), score: 0.7 NADKNAD kinase (ENSG00000008130), score: -0.46 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (ENSG00000103174), score: -0.48 NARS2asparaginyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000137513), score: 0.42 NDUFA12NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 (ENSG00000184752), score: 0.59 NDUFA8NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa (ENSG00000119421), score: 0.46 NDUFB2NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa (ENSG00000090266), score: 0.43 NDUFS3NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) (ENSG00000213619), score: 0.49 NEBLnebulette (ENSG00000078114), score: 0.45 NEK9NIMA (never in mitosis gene a)- related kinase 9 (ENSG00000119638), score: 0.51 NFU1NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) (ENSG00000169599), score: 0.52 NGEFneuronal guanine nucleotide exchange factor (ENSG00000066248), score: -0.45 NHLRC2NHL repeat containing 2 (ENSG00000196865), score: 0.64 NINninein (GSK3B interacting protein) (ENSG00000100503), score: 0.47 NRAPnebulin-related anchoring protein (ENSG00000197893), score: 0.61 NRBP2nuclear receptor binding protein 2 (ENSG00000185189), score: -0.47 NT5C1A5'-nucleotidase, cytosolic IA (ENSG00000116981), score: 0.46 NUBPLnucleotide binding protein-like (ENSG00000151413), score: 0.52 NUP85nucleoporin 85kDa (ENSG00000125450), score: -0.48 P2RX1purinergic receptor P2X, ligand-gated ion channel, 1 (ENSG00000108405), score: 0.57 P2RY2purinergic receptor P2Y, G-protein coupled, 2 (ENSG00000175591), score: 0.42 PAIP1poly(A) binding protein interacting protein 1 (ENSG00000172239), score: 0.46 PALMDpalmdelphin (ENSG00000099260), score: 0.55 PAMpeptidylglycine alpha-amidating monooxygenase (ENSG00000145730), score: 0.47 PAOXpolyamine oxidase (exo-N4-amino) (ENSG00000148832), score: -0.45 PBX3pre-B-cell leukemia homeobox 3 (ENSG00000167081), score: 0.59 PCDH7protocadherin 7 (ENSG00000169851), score: 0.43 PDCD5programmed cell death 5 (ENSG00000105185), score: 0.42 PDE3Aphosphodiesterase 3A, cGMP-inhibited (ENSG00000172572), score: 0.64 PDE7Aphosphodiesterase 7A (ENSG00000205268), score: 0.47 PDGFRLplatelet-derived growth factor receptor-like (ENSG00000104213), score: 0.45 PDHBpyruvate dehydrogenase (lipoamide) beta (ENSG00000168291), score: 0.45 PDLIM5PDZ and LIM domain 5 (ENSG00000163110), score: 0.53 PDZRN3PDZ domain containing ring finger 3 (ENSG00000121440), score: 0.55 PEX16peroxisomal biogenesis factor 16 (ENSG00000121680), score: -0.48 PFKFB46-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (ENSG00000114268), score: -0.48 PHLPP1PH domain and leucine rich repeat protein phosphatase 1 (ENSG00000081913), score: -0.45 PIGNphosphatidylinositol glycan anchor biosynthesis, class N (ENSG00000197563), score: 0.51 PLAAphospholipase A2-activating protein (ENSG00000137055), score: 0.47 PLCL1phospholipase C-like 1 (ENSG00000115896), score: 0.63 PLCXD2phosphatidylinositol-specific phospholipase C, X domain containing 2 (ENSG00000144824), score: 0.49 PLEKHA2pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 (ENSG00000169499), score: 0.56 PLNphospholamban (ENSG00000198523), score: 0.64 POF1Bpremature ovarian failure, 1B (ENSG00000124429), score: 0.77 POLG2polymerase (DNA directed), gamma 2, accessory subunit (ENSG00000136480), score: 0.41 POSTNperiostin, osteoblast specific factor (ENSG00000133110), score: 0.68 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (ENSG00000109819), score: 0.44 PPFIBP2PTPRF interacting protein, binding protein 2 (liprin beta 2) (ENSG00000166387), score: -0.45 PPIGpeptidylprolyl isomerase G (cyclophilin G) (ENSG00000138398), score: 0.44 PPP1R14Cprotein phosphatase 1, regulatory (inhibitor) subunit 14C (ENSG00000198729), score: 0.58 PPP1R1Cprotein phosphatase 1, regulatory (inhibitor) subunit 1C (ENSG00000150722), score: 0.69 PPP1R3Aprotein phosphatase 1, regulatory (inhibitor) subunit 3A (ENSG00000154415), score: 0.83 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: -0.48 PPTC7PTC7 protein phosphatase homolog (S. cerevisiae) (ENSG00000196850), score: 0.5 PRKAG2protein kinase, AMP-activated, gamma 2 non-catalytic subunit (ENSG00000106617), score: 0.41 PRKCHprotein kinase C, eta (ENSG00000027075), score: 0.43 PSMA1proteasome (prosome, macropain) subunit, alpha type, 1 (ENSG00000129084), score: 0.43 PSMA4proteasome (prosome, macropain) subunit, alpha type, 4 (ENSG00000041357), score: 0.47 PSMD14proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 (ENSG00000115233), score: 0.47 PSMD6proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 (ENSG00000163636), score: 0.44 PTDSS1phosphatidylserine synthase 1 (ENSG00000156471), score: 0.79 PTGFRNprostaglandin F2 receptor negative regulator (ENSG00000134247), score: 0.5 PTRFpolymerase I and transcript release factor (ENSG00000177469), score: 0.47 RAB11FIP1RAB11 family interacting protein 1 (class I) (ENSG00000156675), score: 0.59 RAB40CRAB40C, member RAS oncogene family (ENSG00000197562), score: -0.46 RAB9BRAB9B, member RAS oncogene family (ENSG00000123570), score: 0.47 RAD9ARAD9 homolog A (S. pombe) (ENSG00000172613), score: -0.58 RBM20RNA binding motif protein 20 (ENSG00000203867), score: 0.62 RBM24RNA binding motif protein 24 (ENSG00000112183), score: 0.66 RBPMS2RNA binding protein with multiple splicing 2 (ENSG00000166831), score: 0.47 RCAN1regulator of calcineurin 1 (ENSG00000159200), score: 0.47 REXO2REX2, RNA exonuclease 2 homolog (S. cerevisiae) (ENSG00000076043), score: 0.52 RHOT1ras homolog gene family, member T1 (ENSG00000126858), score: 0.42 RILPL1Rab interacting lysosomal protein-like 1 (ENSG00000188026), score: 0.42 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (ENSG00000067900), score: 0.48 RPF1ribosome production factor 1 homolog (S. cerevisiae) (ENSG00000117133), score: 0.44 RPL3Lribosomal protein L3-like (ENSG00000140986), score: 0.53 RRADRas-related associated with diabetes (ENSG00000166592), score: 0.45 RSAD2radical S-adenosyl methionine domain containing 2 (ENSG00000134321), score: 0.49 RYR2ryanodine receptor 2 (cardiac) (ENSG00000198626), score: 0.6 SALL1sal-like 1 (Drosophila) (ENSG00000103449), score: -0.45 SEMA3Csema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C (ENSG00000075223), score: 0.53 SGCGsarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) (ENSG00000102683), score: 0.65 SGPP1sphingosine-1-phosphate phosphatase 1 (ENSG00000126821), score: -0.45 SH3RF2SH3 domain containing ring finger 2 (ENSG00000156463), score: 0.45 SHROOM2shroom family member 2 (ENSG00000146950), score: -0.46 SIK3SIK family kinase 3 (ENSG00000160584), score: -0.49 SIPA1L1signal-induced proliferation-associated 1 like 1 (ENSG00000197555), score: -0.51 SIRT5sirtuin 5 (ENSG00000124523), score: 0.52 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (ENSG00000155380), score: 0.51 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: -0.46 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (ENSG00000100372), score: -0.5 SLC25A3solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 (ENSG00000075415), score: 0.46 SLC25A30solute carrier family 25, member 30 (ENSG00000174032), score: 0.67 SLC25A33solute carrier family 25, member 33 (ENSG00000171612), score: -0.44 SLC26A9solute carrier family 26, member 9 (ENSG00000174502), score: 0.75 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (ENSG00000136856), score: -0.52 SLC41A1solute carrier family 41, member 1 (ENSG00000133065), score: 0.63 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (ENSG00000183023), score: 0.62 SMPXsmall muscle protein, X-linked (ENSG00000091482), score: 0.61 SMYD1SET and MYND domain containing 1 (ENSG00000115593), score: 0.57 SMYD2SET and MYND domain containing 2 (ENSG00000143499), score: 0.6 SNAPC4small nuclear RNA activating complex, polypeptide 4, 190kDa (ENSG00000165684), score: -0.58 SNX3sorting nexin 3 (ENSG00000112335), score: 0.55 SNX6sorting nexin 6 (ENSG00000129515), score: 0.45 SORBS1sorbin and SH3 domain containing 1 (ENSG00000095637), score: 0.55 SQLEsqualene epoxidase (ENSG00000104549), score: -0.46 SSBP1single-stranded DNA binding protein 1 (ENSG00000106028), score: 0.49 STRN3striatin, calmodulin binding protein 3 (ENSG00000196792), score: 0.44 STX11syntaxin 11 (ENSG00000135604), score: 0.5 STX17syntaxin 17 (ENSG00000136874), score: 0.46 SWAP70SWAP switching B-cell complex 70kDa subunit (ENSG00000133789), score: 0.45 SYF2SYF2 homolog, RNA splicing factor (S. cerevisiae) (ENSG00000117614), score: 0.47 SYNMsynemin, intermediate filament protein (ENSG00000182253), score: 0.63 SYNPO2synaptopodin 2 (ENSG00000172403), score: 0.57 TAB2TGF-beta activated kinase 1/MAP3K7 binding protein 2 (ENSG00000055208), score: 0.45 TACC2transforming, acidic coiled-coil containing protein 2 (ENSG00000138162), score: 0.5 TBX20T-box 20 (ENSG00000164532), score: 0.66 TBX5T-box 5 (ENSG00000089225), score: 0.53 TEAD1TEA domain family member 1 (SV40 transcriptional enhancer factor) (ENSG00000187079), score: 0.58 TECRLtrans-2,3-enoyl-CoA reductase-like (ENSG00000205678), score: 0.7 TIMM17Atranslocase of inner mitochondrial membrane 17 homolog A (yeast) (ENSG00000134375), score: 0.43 TMEM117transmembrane protein 117 (ENSG00000139173), score: 0.42 TMEM180transmembrane protein 180 (ENSG00000138111), score: -0.52 TMEM182transmembrane protein 182 (ENSG00000170417), score: 0.81 TMOD1tropomodulin 1 (ENSG00000136842), score: 0.47 TMX4thioredoxin-related transmembrane protein 4 (ENSG00000125827), score: 0.52 TNNC1troponin C type 1 (slow) (ENSG00000114854), score: 0.64 TNNT2troponin T type 2 (cardiac) (ENSG00000118194), score: 0.53 TOP2Btopoisomerase (DNA) II beta 180kDa (ENSG00000077097), score: 0.49 TOR2Atorsin family 2, member A (ENSG00000160404), score: -0.53 TPM1tropomyosin 1 (alpha) (ENSG00000140416), score: 0.7 TPST1tyrosylprotein sulfotransferase 1 (ENSG00000169902), score: -0.45 TRAF2TNF receptor-associated factor 2 (ENSG00000127191), score: -0.65 TRIM55tripartite motif-containing 55 (ENSG00000147573), score: 0.51 TRIM63tripartite motif-containing 63 (ENSG00000158022), score: 0.6 TSEN54tRNA splicing endonuclease 54 homolog (S. cerevisiae) (ENSG00000182173), score: -0.53 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: 0.45 TXLNBtaxilin beta (ENSG00000164440), score: 0.61 TYRP1tyrosinase-related protein 1 (ENSG00000107165), score: 0.81 UQCRC1ubiquinol-cytochrome c reductase core protein I (ENSG00000010256), score: 0.44 UQCRC2ubiquinol-cytochrome c reductase core protein II (ENSG00000140740), score: 0.49 USP28ubiquitin specific peptidase 28 (ENSG00000048028), score: 0.5 USP47ubiquitin specific peptidase 47 (ENSG00000170242), score: 0.44 VEZTvezatin, adherens junctions transmembrane protein (ENSG00000028203), score: 0.51 VIPR2vasoactive intestinal peptide receptor 2 (ENSG00000106018), score: 0.52 VPS16vacuolar protein sorting 16 homolog (S. cerevisiae) (ENSG00000215305), score: -0.5 VRK2vaccinia related kinase 2 (ENSG00000028116), score: 0.5 WDR91WD repeat domain 91 (ENSG00000105875), score: -0.55 WISP2WNT1 inducible signaling pathway protein 2 (ENSG00000064205), score: 0.51 WTIPWilms tumor 1 interacting protein (ENSG00000142279), score: 0.5 XPO4exportin 4 (ENSG00000132953), score: 0.66 YME1L1YME1-like 1 (S. cerevisiae) (ENSG00000136758), score: 0.45 YRDCyrdC domain containing (E. coli) (ENSG00000196449), score: -0.57 ZAKsterile alpha motif and leucine zipper containing kinase AZK (ENSG00000091436), score: 0.74 ZBTB43zinc finger and BTB domain containing 43 (ENSG00000169155), score: 0.71 ZEB1zinc finger E-box binding homeobox 1 (ENSG00000148516), score: 0.59 ZNF276zinc finger protein 276 (ENSG00000158805), score: -0.55 ZNF330zinc finger protein 330 (ENSG00000109445), score: 0.81 ZNF414zinc finger protein 414 (ENSG00000133250), score: -0.5 ZNF821zinc finger protein 821 (ENSG00000102984), score: -0.51 ZNRF3zinc and ring finger 3 (ENSG00000183579), score: -0.44
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_ht_f_ca1 | ptr | ht | f | _ |
| ppy_ht_m_ca1 | ppy | ht | m | _ |
| ppa_ht_f_ca1 | ppa | ht | f | _ |
| ptr_ht_m_ca1 | ptr | ht | m | _ |