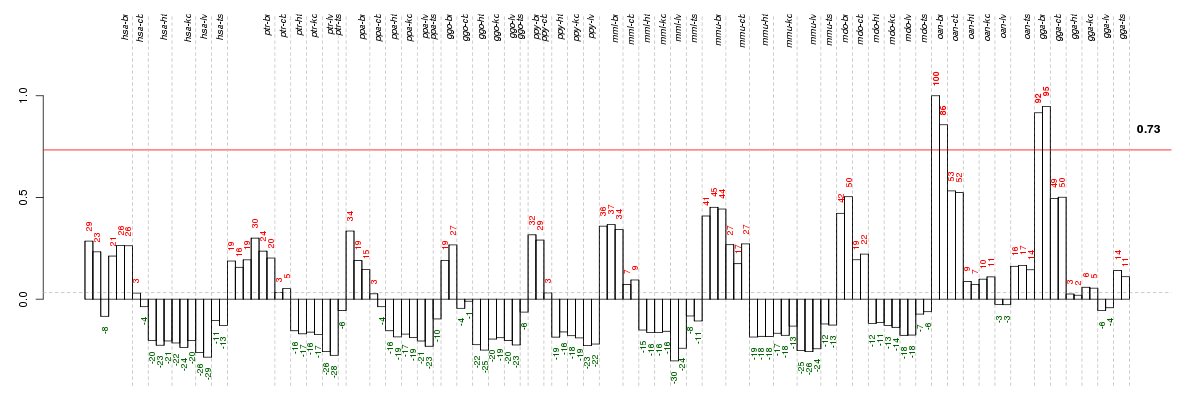

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
G-protein coupled receptor protein signaling pathway
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
behavior
The specific actions or reactions of an organism in response to external or internal stimuli. Patterned activity of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
NA
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse
The junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell; the site of interneuronal communication. As the nerve fiber approaches the synapse it enlarges into a specialized structure, the presynaptic nerve ending, which contains mitochondria and synaptic vesicles. At the tip of the nerve ending is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic nerve ending secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane.
postsynaptic membrane
A specialized area of membrane facing the presynaptic membrane on the tip of the nerve ending and separated from it by a minute cleft (the synaptic cleft). Neurotransmitters across the synaptic cleft and transmit the signal to the postsynaptic membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
postsynaptic membrane
A specialized area of membrane facing the presynaptic membrane on the tip of the nerve ending and separated from it by a minute cleft (the synaptic cleft). Neurotransmitters across the synaptic cleft and transmit the signal to the postsynaptic membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

peptide receptor activity
Combining with an extracellular or intracellular peptide to initiate a change in cell activity.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
peptide receptor activity, G-protein coupled
Combining with an extracellular or intracellular peptide to initiate a G-protein mediated change in cell activity. A G-protein is a signal transduction molecule that alternates between an inactive GDP-bound and an active GTP-bound state.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
neurotransmitter binding
Interacting selectively and non-covalently with a neurotransmitter, any chemical substance that is capable of transmitting (or inhibiting the transmission of) a nerve impulse from a neuron to another cell.
peptide binding
Interacting selectively and non-covalently with peptides, any of a group of organic compounds comprising two or more amino acids linked by peptide bonds.
neuropeptide binding
Interacting selectively and non-covalently and stoichiometrically with neuropeptides, peptides with direct synaptic effects (peptide neurotransmitters) or indirect modulatory effects on the nervous system (peptide neuromodulators).
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
peptide receptor activity
Combining with an extracellular or intracellular peptide to initiate a change in cell activity.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
peptide receptor activity, G-protein coupled
Combining with an extracellular or intracellular peptide to initiate a G-protein mediated change in cell activity. A G-protein is a signal transduction molecule that alternates between an inactive GDP-bound and an active GTP-bound state.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 3.981e-06 | 5.844 | 22 | 102 | Neuroactive ligand-receptor interaction |
AAK1AP2 associated kinase 1 (ENSG00000115977), score: 0.41 AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.42 ACCN4amiloride-sensitive cation channel 4, pituitary (ENSG00000072182), score: 0.5 ACSF3acyl-CoA synthetase family member 3 (ENSG00000176715), score: -0.33 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.49 ADCY8adenylate cyclase 8 (brain) (ENSG00000155897), score: 0.44 ADCYAP1adenylate cyclase activating polypeptide 1 (pituitary) (ENSG00000141433), score: 0.43 ALKanaplastic lymphoma receptor tyrosine kinase (ENSG00000171094), score: 0.46 ALX4ALX homeobox 4 (ENSG00000052850), score: 0.48 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: -0.36 ANKFN1ankyrin-repeat and fibronectin type III domain containing 1 (ENSG00000153930), score: 0.48 ANKRA2ankyrin repeat, family A (RFXANK-like), 2 (ENSG00000164331), score: 0.48 ANKRD27ankyrin repeat domain 27 (VPS9 domain) (ENSG00000105186), score: -0.5 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: -0.36 ANKRD44ankyrin repeat domain 44 (ENSG00000065413), score: 0.42 ANO3anoctamin 3 (ENSG00000134343), score: 0.4 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: -0.41 ARHGAP6Rho GTPase activating protein 6 (ENSG00000047648), score: 0.41 ARIH2ariadne homolog 2 (Drosophila) (ENSG00000177479), score: -0.37 ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: -0.35 ARPP21cAMP-regulated phosphoprotein, 21kDa (ENSG00000172995), score: 0.43 ATP13A1ATPase type 13A1 (ENSG00000105726), score: -0.37 ATP13A5ATPase type 13A5 (ENSG00000187527), score: 0.4 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.45 ATXN7ataxin 7 (ENSG00000163635), score: -0.36 AURKAIP1aurora kinase A interacting protein 1 (ENSG00000175756), score: -0.37 AVPR1Barginine vasopressin receptor 1B (ENSG00000198049), score: 0.71 B3GALT1UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 (ENSG00000172318), score: 0.46 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.48 BACE1beta-site APP-cleaving enzyme 1 (ENSG00000186318), score: 0.41 BAT2L2HLA-B associated transcript 2-like 2 (ENSG00000117523), score: 0.4 BCL11BB-cell CLL/lymphoma 11B (zinc finger protein) (ENSG00000127152), score: 0.58 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.44 BMP3bone morphogenetic protein 3 (ENSG00000152785), score: 0.62 BRS3bombesin-like receptor 3 (ENSG00000102239), score: 0.88 C12orf29chromosome 12 open reading frame 29 (ENSG00000133641), score: 0.4 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.39 C14orf37chromosome 14 open reading frame 37 (ENSG00000139971), score: 0.47 C15orf60chromosome 15 open reading frame 60 (ENSG00000183324), score: 0.43 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.44 C20orf103chromosome 20 open reading frame 103 (ENSG00000125869), score: 0.41 C20orf186chromosome 20 open reading frame 186 (ENSG00000186191), score: 0.65 C20orf72chromosome 20 open reading frame 72 (ENSG00000125871), score: -0.33 C22orf13chromosome 22 open reading frame 13 (ENSG00000138867), score: -0.34 C5orf24chromosome 5 open reading frame 24 (ENSG00000181904), score: 0.48 C6orf106chromosome 6 open reading frame 106 (ENSG00000196821), score: -0.5 C6orf186chromosome 6 open reading frame 186 (ENSG00000053328), score: 0.46 CACNA1Ecalcium channel, voltage-dependent, R type, alpha 1E subunit (ENSG00000198216), score: 0.43 CACNG3calcium channel, voltage-dependent, gamma subunit 3 (ENSG00000006116), score: 0.47 CASD1CAS1 domain containing 1 (ENSG00000127995), score: 0.44 CASKcalcium/calmodulin-dependent serine protein kinase (MAGUK family) (ENSG00000147044), score: 0.4 CASTcalpastatin (ENSG00000153113), score: -0.37 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: -0.36 CCND3cyclin D3 (ENSG00000112576), score: -0.37 CDC40cell division cycle 40 homolog (S. cerevisiae) (ENSG00000168438), score: 0.51 CDH10cadherin 10, type 2 (T2-cadherin) (ENSG00000040731), score: 0.41 CDH12cadherin 12, type 2 (N-cadherin 2) (ENSG00000154162), score: 0.41 CDH19cadherin 19, type 2 (ENSG00000071991), score: 0.46 CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.45 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (ENSG00000113361), score: 0.43 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.43 CDH9cadherin 9, type 2 (T1-cadherin) (ENSG00000113100), score: 0.44 CDK17cyclin-dependent kinase 17 (ENSG00000059758), score: 0.46 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.45 CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: -0.4 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: -0.34 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.45 CINPcyclin-dependent kinase 2 interacting protein (ENSG00000100865), score: -0.33 CLCN3chloride channel 3 (ENSG00000109572), score: 0.53 CLVS2clavesin 2 (ENSG00000146352), score: 0.41 CNGB1cyclic nucleotide gated channel beta 1 (ENSG00000070729), score: 0.6 CNOT2CCR4-NOT transcription complex, subunit 2 (ENSG00000111596), score: -0.38 CNTN3contactin 3 (plasmacytoma associated) (ENSG00000113805), score: 0.61 CNTN4contactin 4 (ENSG00000144619), score: 0.45 CNTN5contactin 5 (ENSG00000149972), score: 0.79 COL19A1collagen, type XIX, alpha 1 (ENSG00000082293), score: 0.41 COL25A1collagen, type XXV, alpha 1 (ENSG00000188517), score: 0.51 COMMD2COMM domain containing 2 (ENSG00000114744), score: 0.46 CPLX4complexin 4 (ENSG00000166569), score: 0.52 CPNE4copine IV (ENSG00000196353), score: 0.41 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.43 CREB3L1cAMP responsive element binding protein 3-like 1 (ENSG00000157613), score: 0.4 CRHcorticotropin releasing hormone (ENSG00000147571), score: 0.42 CSMD2CUB and Sushi multiple domains 2 (ENSG00000121904), score: 0.48 CTNND1catenin (cadherin-associated protein), delta 1 (ENSG00000198561), score: -0.52 CUL4Acullin 4A (ENSG00000139842), score: -0.58 DAZAP1DAZ associated protein 1 (ENSG00000071626), score: -0.4 DCLK3doublecortin-like kinase 3 (ENSG00000163673), score: 0.48 DCNdecorin (ENSG00000011465), score: -0.34 DCTDdCMP deaminase (ENSG00000129187), score: -0.33 DCTN4dynactin 4 (p62) (ENSG00000132912), score: 0.5 DDX24DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 (ENSG00000089737), score: -0.38 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.57 DLGAP2discs, large (Drosophila) homolog-associated protein 2 (ENSG00000198010), score: 0.41 DLK1delta-like 1 homolog (Drosophila) (ENSG00000185559), score: 0.45 DLX6distal-less homeobox 6 (ENSG00000006377), score: 0.59 DRD2dopamine receptor D2 (ENSG00000149295), score: 0.48 DSCAMDown syndrome cell adhesion molecule (ENSG00000171587), score: 0.43 DUSP19dual specificity phosphatase 19 (ENSG00000162999), score: 0.5 DYRK1Adual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (ENSG00000157540), score: -0.34 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.4 EIF2C2eukaryotic translation initiation factor 2C, 2 (ENSG00000123908), score: -0.35 ELAVL4ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) (ENSG00000162374), score: 0.4 ELP2elongation protein 2 homolog (S. cerevisiae) (ENSG00000134759), score: -0.4 ENTPD2ectonucleoside triphosphate diphosphohydrolase 2 (ENSG00000054179), score: 0.44 EPHA3EPH receptor A3 (ENSG00000044524), score: 0.54 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.62 EPHA7EPH receptor A7 (ENSG00000135333), score: 0.45 EPHX4epoxide hydrolase 4 (ENSG00000172031), score: 0.46 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (ENSG00000178568), score: 0.44 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.41 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: 0.46 FAM164Afamily with sequence similarity 164, member A (ENSG00000104427), score: 0.44 FAM43Afamily with sequence similarity 43, member A (ENSG00000185112), score: 0.4 FAM5Cfamily with sequence similarity 5, member C (ENSG00000162670), score: 0.41 FARP2FERM, RhoGEF and pleckstrin domain protein 2 (ENSG00000006607), score: -0.33 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.47 FIBCD1fibrinogen C domain containing 1 (ENSG00000130720), score: 0.6 FOXP2forkhead box P2 (ENSG00000128573), score: 0.41 FRMPD4FERM and PDZ domain containing 4 (ENSG00000169933), score: 0.45 FTSJD2FtsJ methyltransferase domain containing 2 (ENSG00000137200), score: -0.33 FUT11fucosyltransferase 11 (alpha (1,3) fucosyltransferase) (ENSG00000196968), score: -0.42 GAAglucosidase, alpha; acid (ENSG00000171298), score: -0.44 GABRA2gamma-aminobutyric acid (GABA) A receptor, alpha 2 (ENSG00000151834), score: 0.56 GABRA4gamma-aminobutyric acid (GABA) A receptor, alpha 4 (ENSG00000109158), score: 0.51 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.59 GAD2glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) (ENSG00000136750), score: 0.47 GALNTL6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 (ENSG00000174473), score: 0.59 GALR1galanin receptor 1 (ENSG00000166573), score: 0.77 GAMTguanidinoacetate N-methyltransferase (ENSG00000130005), score: -0.35 GBX2gastrulation brain homeobox 2 (ENSG00000168505), score: 0.52 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (ENSG00000158555), score: 0.48 GLRA2glycine receptor, alpha 2 (ENSG00000101958), score: 0.48 GLRA3glycine receptor, alpha 3 (ENSG00000145451), score: 0.51 GLRA4glycine receptor, alpha 4 (ENSG00000188828), score: 0.45 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 0.83 GPRC5CG protein-coupled receptor, family C, group 5, member C (ENSG00000170412), score: -0.34 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.41 GRIN2Bglutamate receptor, ionotropic, N-methyl D-aspartate 2B (ENSG00000150086), score: 0.6 GRIN3Aglutamate receptor, ionotropic, N-methyl-D-aspartate 3A (ENSG00000198785), score: 0.43 GRIP1glutamate receptor interacting protein 1 (ENSG00000155974), score: 0.43 GRM3glutamate receptor, metabotropic 3 (ENSG00000198822), score: 0.42 GUCY2Cguanylate cyclase 2C (heat stable enterotoxin receptor) (ENSG00000070019), score: 0.43 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: -0.41 HEATR5BHEAT repeat containing 5B (ENSG00000008869), score: 0.46 HERPUD1homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 (ENSG00000051108), score: -0.37 HHLA1HERV-H LTR-associating 1 (ENSG00000132297), score: 0.59 HNRNPA0heterogeneous nuclear ribonucleoprotein A0 (ENSG00000177733), score: -0.34 HSF4heat shock transcription factor 4 (ENSG00000102878), score: -0.4 HTR1D5-hydroxytryptamine (serotonin) receptor 1D (ENSG00000179546), score: 0.67 HTR45-hydroxytryptamine (serotonin) receptor 4 (ENSG00000164270), score: 0.72 IAH1isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) (ENSG00000134330), score: -0.34 ICMTisoprenylcysteine carboxyl methyltransferase (ENSG00000116237), score: -0.33 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.61 IL1RAPL2interleukin 1 receptor accessory protein-like 2 (ENSG00000189108), score: 1 ILDR2immunoglobulin-like domain containing receptor 2 (ENSG00000143195), score: 0.42 INPP5Binositol polyphosphate-5-phosphatase, 75kDa (ENSG00000204084), score: -0.33 INPPL1inositol polyphosphate phosphatase-like 1 (ENSG00000165458), score: -0.4 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: 0.56 KBTBD3kelch repeat and BTB (POZ) domain containing 3 (ENSG00000182359), score: 0.47 KCNA4potassium voltage-gated channel, shaker-related subfamily, member 4 (ENSG00000182255), score: 0.55 KCNG3potassium voltage-gated channel, subfamily G, member 3 (ENSG00000171126), score: 0.53 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.41 KCNK2potassium channel, subfamily K, member 2 (ENSG00000082482), score: 0.47 KCNMB2potassium large conductance calcium-activated channel, subfamily M, beta member 2 (ENSG00000197584), score: 0.39 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (ENSG00000184156), score: 0.48 KCNS1potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 (ENSG00000124134), score: 0.41 KCNS2potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 (ENSG00000156486), score: 0.66 KCTD4potassium channel tetramerisation domain containing 4 (ENSG00000180332), score: 0.51 KIAA0368KIAA0368 (ENSG00000136813), score: -0.35 KIAA2022KIAA2022 (ENSG00000050030), score: 0.51 KLHDC2kelch domain containing 2 (ENSG00000165516), score: 0.43 KLHDC3kelch domain containing 3 (ENSG00000124702), score: -0.35 KLHL20kelch-like 20 (Drosophila) (ENSG00000076321), score: 0.43 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.41 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.49 LMO1LIM domain only 1 (rhombotin 1) (ENSG00000166407), score: 0.53 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: -0.45 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.6 LOC100293905similar to gastrin-releasing peptide receptor (ENSG00000126010), score: 0.41 LOH12CR1loss of heterozygosity, 12, chromosomal region 1 (ENSG00000165714), score: 0.4 LPIN1lipin 1 (ENSG00000134324), score: -0.35 LPPR4lipid phosphate phosphatase-related protein type 4 (ENSG00000117600), score: 0.41 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (ENSG00000126243), score: 0.42 LRFN5leucine rich repeat and fibronectin type III domain containing 5 (ENSG00000165379), score: 0.46 LRMPlymphoid-restricted membrane protein (ENSG00000118308), score: 0.61 LRP12low density lipoprotein receptor-related protein 12 (ENSG00000147650), score: 0.44 LRRC33leucine rich repeat containing 33 (ENSG00000174004), score: -0.35 LRRC40leucine rich repeat containing 40 (ENSG00000066557), score: 0.43 LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.44 LRRC7leucine rich repeat containing 7 (ENSG00000033122), score: 0.61 LZTS2leucine zipper, putative tumor suppressor 2 (ENSG00000107816), score: -0.33 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.35 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: -0.42 MAP2K1mitogen-activated protein kinase kinase 1 (ENSG00000169032), score: 0.44 MAP3K3mitogen-activated protein kinase kinase kinase 3 (ENSG00000198909), score: -0.38 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: -0.4 MC4Rmelanocortin 4 receptor (ENSG00000166603), score: 0.4 MCF2MCF.2 cell line derived transforming sequence (ENSG00000101977), score: 0.55 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.58 METTL9methyltransferase like 9 (ENSG00000197006), score: -0.34 MLXIPMLX interacting protein (ENSG00000175727), score: -0.6 MOSPD2motile sperm domain containing 2 (ENSG00000130150), score: 0.44 MPHOSPH8M-phase phosphoprotein 8 (ENSG00000196199), score: -0.34 MTMR2myotubularin related protein 2 (ENSG00000087053), score: 0.44 MYOGmyogenin (myogenic factor 4) (ENSG00000122180), score: 0.57 NARFLnuclear prelamin A recognition factor-like (ENSG00000103245), score: -0.36 NDST4N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 (ENSG00000138653), score: 0.58 NECAB1N-terminal EF-hand calcium binding protein 1 (ENSG00000123119), score: 0.46 NECAP2NECAP endocytosis associated 2 (ENSG00000157191), score: -0.4 NEK9NIMA (never in mitosis gene a)- related kinase 9 (ENSG00000119638), score: -0.41 NETO1neuropilin (NRP) and tolloid (TLL)-like 1 (ENSG00000166342), score: 0.43 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (ENSG00000109320), score: -0.45 NINJ1ninjurin 1 (ENSG00000131669), score: -0.37 NMNAT3nicotinamide nucleotide adenylyltransferase 3 (ENSG00000163864), score: -0.37 NPY2Rneuropeptide Y receptor Y2 (ENSG00000185149), score: 0.9 NPY5Rneuropeptide Y receptor Y5 (ENSG00000164129), score: 0.48 NTSneurotensin (ENSG00000133636), score: 0.82 NTSR1neurotensin receptor 1 (high affinity) (ENSG00000101188), score: 0.48 OFCC1orofacial cleft 1 candidate 1 (ENSG00000181355), score: 0.55 OLIG3oligodendrocyte transcription factor 3 (ENSG00000177468), score: 0.99 OPCMLopioid binding protein/cell adhesion molecule-like (ENSG00000183715), score: 0.42 OPRK1opioid receptor, kappa 1 (ENSG00000082556), score: 0.65 OSTF1osteoclast stimulating factor 1 (ENSG00000134996), score: -0.36 OTOFotoferlin (ENSG00000115155), score: 0.54 P2RY1purinergic receptor P2Y, G-protein coupled, 1 (ENSG00000169860), score: 0.5 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: 0.4 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.36 PCDH19protocadherin 19 (ENSG00000165194), score: 0.45 PDRG1p53 and DNA-damage regulated 1 (ENSG00000088356), score: 0.39 PENKproenkephalin (ENSG00000181195), score: 0.41 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (ENSG00000182621), score: 0.48 POLGpolymerase (DNA directed), gamma (ENSG00000140521), score: -0.4 POU1F1POU class 1 homeobox 1 (ENSG00000064835), score: 0.69 PPP3CAprotein phosphatase 3, catalytic subunit, alpha isozyme (ENSG00000138814), score: 0.41 PSKH1protein serine kinase H1 (ENSG00000159792), score: -0.34 PTPN1protein tyrosine phosphatase, non-receptor type 1 (ENSG00000196396), score: -0.39 PTPN12protein tyrosine phosphatase, non-receptor type 12 (ENSG00000127947), score: -0.33 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: -0.36 R3HDM1R3H domain containing 1 (ENSG00000048991), score: 0.42 RAB33BRAB33B, member RAS oncogene family (ENSG00000172007), score: 0.43 RAB3GAP2RAB3 GTPase activating protein subunit 2 (non-catalytic) (ENSG00000118873), score: 0.45 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.38 RASD2RASD family, member 2 (ENSG00000100302), score: 0.42 RECQL5RecQ protein-like 5 (ENSG00000108469), score: -0.34 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.44 RHOT2ras homolog gene family, member T2 (ENSG00000140983), score: -0.36 RIC3resistance to inhibitors of cholinesterase 3 homolog (C. elegans) (ENSG00000166405), score: 0.43 RNF114ring finger protein 114 (ENSG00000124226), score: -0.37 RNF43ring finger protein 43 (ENSG00000108375), score: -0.34 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: 0.42 RPE65retinal pigment epithelium-specific protein 65kDa (ENSG00000116745), score: 0.59 RPS6KC1ribosomal protein S6 kinase, 52kDa, polypeptide 1 (ENSG00000136643), score: 0.44 RSPO3R-spondin 3 homolog (Xenopus laevis) (ENSG00000146374), score: 0.49 RXFP3relaxin/insulin-like family peptide receptor 3 (ENSG00000182631), score: 0.48 SCAMP1secretory carrier membrane protein 1 (ENSG00000085365), score: 0.44 SCG2secretogranin II (ENSG00000171951), score: 0.44 SCGNsecretagogin, EF-hand calcium binding protein (ENSG00000079689), score: 0.45 SEMA3Asema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A (ENSG00000075213), score: 0.58 SEPT6septin 6 (ENSG00000125354), score: 0.4 SFRS15splicing factor, arginine/serine-rich 15 (ENSG00000156304), score: -0.33 SH2B3SH2B adaptor protein 3 (ENSG00000111252), score: -0.36 SLBPstem-loop binding protein (ENSG00000163950), score: -0.33 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (ENSG00000117479), score: -0.35 SLC25A37solute carrier family 25, member 37 (ENSG00000147454), score: -0.52 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.43 SLC5A7solute carrier family 5 (choline transporter), member 7 (ENSG00000115665), score: 0.91 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.44 SLC7A14solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 (ENSG00000013293), score: 0.41 SNAP23synaptosomal-associated protein, 23kDa (ENSG00000092531), score: -0.35 SNRPA1small nuclear ribonucleoprotein polypeptide A' (ENSG00000131876), score: -0.34 SNTG1syntrophin, gamma 1 (ENSG00000147481), score: 0.45 SNX5sorting nexin 5 (ENSG00000089006), score: -0.35 SPON1spondin 1, extracellular matrix protein (ENSG00000152268), score: 0.4 SPPL2Bsignal peptide peptidase-like 2B (ENSG00000005206), score: -0.36 SSTsomatostatin (ENSG00000157005), score: 0.45 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (ENSG00000124214), score: 0.45 STK40serine/threonine kinase 40 (ENSG00000196182), score: -0.35 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.36 STX12syntaxin 12 (ENSG00000117758), score: 0.45 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.43 SUPT5Hsuppressor of Ty 5 homolog (S. cerevisiae) (ENSG00000196235), score: -0.35 SV2Csynaptic vesicle glycoprotein 2C (ENSG00000122012), score: 0.71 SYNJ1synaptojanin 1 (ENSG00000159082), score: 0.43 TT, brachyury homolog (mouse) (ENSG00000164458), score: 0.4 TAC1tachykinin, precursor 1 (ENSG00000006128), score: 0.7 TAPT1transmembrane anterior posterior transformation 1 (ENSG00000169762), score: -0.36 TBC1D30TBC1 domain family, member 30 (ENSG00000111490), score: 0.42 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: 0.45 TDRD7tudor domain containing 7 (ENSG00000196116), score: -0.39 TFAP2Dtranscription factor AP-2 delta (activating enhancer binding protein 2 delta) (ENSG00000008197), score: 0.89 THSD7Athrombospondin, type I, domain containing 7A (ENSG00000005108), score: 0.59 THY1Thy-1 cell surface antigen (ENSG00000154096), score: 0.4 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: -0.34 TMEFF2transmembrane protein with EGF-like and two follistatin-like domains 2 (ENSG00000144339), score: 0.44 TMEM117transmembrane protein 117 (ENSG00000139173), score: 0.49 TMEM200Atransmembrane protein 200A (ENSG00000164484), score: 0.5 TMEM74transmembrane protein 74 (ENSG00000164841), score: 0.42 TMEM9BTMEM9 domain family, member B (ENSG00000175348), score: 0.4 TOM1L1target of myb1 (chicken)-like 1 (ENSG00000141198), score: -0.4 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: -0.41 TOR2Atorsin family 2, member A (ENSG00000160404), score: -0.39 TPBGtrophoblast glycoprotein (ENSG00000146242), score: 0.57 TPRA1transmembrane protein, adipocyte asscociated 1 (ENSG00000163870), score: -0.35 TRABDTraB domain containing (ENSG00000170638), score: -0.35 TRPA1transient receptor potential cation channel, subfamily A, member 1 (ENSG00000104321), score: 0.41 TRPC1transient receptor potential cation channel, subfamily C, member 1 (ENSG00000144935), score: 0.49 TRPC5transient receptor potential cation channel, subfamily C, member 5 (ENSG00000072315), score: 0.69 TRPM2transient receptor potential cation channel, subfamily M, member 2 (ENSG00000142185), score: 0.41 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: -0.34 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.36 UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.38 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: -0.34 UNC5Dunc-5 homolog D (C. elegans) (ENSG00000156687), score: 0.4 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.43 VSTM2AV-set and transmembrane domain containing 2A (ENSG00000170419), score: 0.45 VWC2Lvon Willebrand factor C domain-containing protein 2-like (ENSG00000174453), score: 0.51 WACWW domain containing adaptor with coiled-coil (ENSG00000095787), score: -0.4 WDR26WD repeat domain 26 (ENSG00000162923), score: -0.34 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.4 WNT3Awingless-type MMTV integration site family, member 3A (ENSG00000154342), score: 0.65 WNT4wingless-type MMTV integration site family, member 4 (ENSG00000162552), score: 0.6 XRCC4X-ray repair complementing defective repair in Chinese hamster cells 4 (ENSG00000152422), score: 0.41 ZC3H12Bzinc finger CCCH-type containing 12B (ENSG00000102053), score: 0.44 ZDHHC15zinc finger, DHHC-type containing 15 (ENSG00000102383), score: 0.42 ZFHX4zinc finger homeobox 4 (ENSG00000091656), score: 0.45 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.48 ZNFX1zinc finger, NFX1-type containing 1 (ENSG00000124201), score: -0.39 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.36
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_br_f_ca1 | oan | br | f | _ |
| gga_br_m_ca1 | gga | br | m | _ |
| gga_br_f_ca1 | gga | br | f | _ |
| oan_br_m_ca1 | oan | br | m | _ |