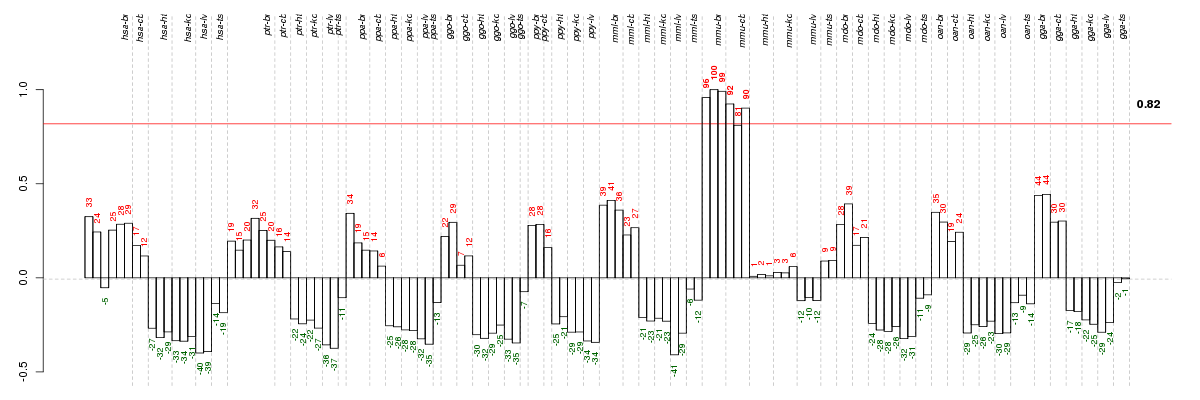

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
all
NA
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.

membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (ENSG00000160179), score: 0.43 ACADSacyl-CoA dehydrogenase, C-2 to C-3 short chain (ENSG00000122971), score: -0.43 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (ENSG00000156140), score: 0.48 ADPRHADP-ribosylarginine hydrolase (ENSG00000144843), score: 0.49 AFF4AF4/FMR2 family, member 4 (ENSG00000072364), score: 0.44 AHI1Abelson helper integration site 1 (ENSG00000135541), score: 0.64 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: 0.48 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: 0.43 ANGEL2angel homolog 2 (Drosophila) (ENSG00000174606), score: 0.48 ANKRD16ankyrin repeat domain 16 (ENSG00000134461), score: 0.44 ANKRD46ankyrin repeat domain 46 (ENSG00000186106), score: 0.54 ANO3anoctamin 3 (ENSG00000134343), score: 0.5 ANO4anoctamin 4 (ENSG00000151572), score: 0.42 ANTXR2anthrax toxin receptor 2 (ENSG00000163297), score: -0.51 ANXA1annexin A1 (ENSG00000135046), score: -0.48 AP4B1adaptor-related protein complex 4, beta 1 subunit (ENSG00000134262), score: -0.44 APPL2adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 (ENSG00000136044), score: 0.5 ARCactivity-regulated cytoskeleton-associated protein (ENSG00000198576), score: 0.51 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (ENSG00000149182), score: -0.44 ARSBarylsulfatase B (ENSG00000113273), score: 0.49 ATG4AATG4 autophagy related 4 homolog A (S. cerevisiae) (ENSG00000101844), score: -0.44 ATP13A5ATPase type 13A5 (ENSG00000187527), score: 0.66 ATP9AATPase, class II, type 9A (ENSG00000054793), score: 0.45 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.88 BAI1brain-specific angiogenesis inhibitor 1 (ENSG00000181790), score: 0.43 BCAT1branched chain amino-acid transaminase 1, cytosolic (ENSG00000060982), score: 0.61 BEGAINbrain-enriched guanylate kinase-associated homolog (rat) (ENSG00000183092), score: 0.44 BFSP2beaded filament structural protein 2, phakinin (ENSG00000170819), score: 0.91 BRMS1Lbreast cancer metastasis-suppressor 1-like (ENSG00000100916), score: 0.53 BTBD8BTB (POZ) domain containing 8 (ENSG00000189195), score: 0.77 C12orf51chromosome 12 open reading frame 51 (ENSG00000173064), score: 0.45 C14orf149chromosome 14 open reading frame 149 (ENSG00000126790), score: -0.48 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: -0.45 C1orf58chromosome 1 open reading frame 58 (ENSG00000162819), score: 0.45 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.42 C20orf103chromosome 20 open reading frame 103 (ENSG00000125869), score: 0.49 C20orf12chromosome 20 open reading frame 12 (ENSG00000089091), score: 0.86 C21orf63chromosome 21 open reading frame 63 (ENSG00000166979), score: -0.43 C6orf130chromosome 6 open reading frame 130 (ENSG00000124596), score: -0.62 C7orf30chromosome 7 open reading frame 30 (ENSG00000156928), score: -0.59 C8orf34chromosome 8 open reading frame 34 (ENSG00000165084), score: 0.59 C9orf95chromosome 9 open reading frame 95 (ENSG00000106733), score: -0.43 CACNA1Ecalcium channel, voltage-dependent, R type, alpha 1E subunit (ENSG00000198216), score: 0.44 CACNG4calcium channel, voltage-dependent, gamma subunit 4 (ENSG00000075461), score: 0.46 CCBL2cysteine conjugate-beta lyase 2 (ENSG00000137944), score: -0.42 CCT4chaperonin containing TCP1, subunit 4 (delta) (ENSG00000115484), score: -0.45 CDC42BPACDC42 binding protein kinase alpha (DMPK-like) (ENSG00000143776), score: 0.48 CDK6cyclin-dependent kinase 6 (ENSG00000105810), score: -0.49 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.57 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.52 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: -0.5 CHRNA6cholinergic receptor, nicotinic, alpha 6 (ENSG00000147434), score: 0.49 CHRNB3cholinergic receptor, nicotinic, beta 3 (ENSG00000147432), score: 0.47 CLVS1clavesin 1 (ENSG00000177182), score: 0.48 CNIH3cornichon homolog 3 (Drosophila) (ENSG00000143786), score: 0.44 COL25A1collagen, type XXV, alpha 1 (ENSG00000188517), score: 0.48 CSMD2CUB and Sushi multiple domains 2 (ENSG00000121904), score: 0.5 CTTNBP2cortactin binding protein 2 (ENSG00000077063), score: 0.48 CYBRD1cytochrome b reductase 1 (ENSG00000071967), score: -0.52 D2HGDHD-2-hydroxyglutarate dehydrogenase (ENSG00000180902), score: -0.44 DACT2dapper, antagonist of beta-catenin, homolog 2 (Xenopus laevis) (ENSG00000164488), score: 0.46 DAGLBdiacylglycerol lipase, beta (ENSG00000164535), score: 0.46 DCTN5dynactin 5 (p25) (ENSG00000166847), score: 0.45 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.47 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (ENSG00000160305), score: 0.46 DMP1dentin matrix acidic phosphoprotein 1 (ENSG00000152592), score: 0.98 DNAJC27DnaJ (Hsp40) homolog, subfamily C, member 27 (ENSG00000115137), score: 0.45 DOPEY2dopey family member 2 (ENSG00000142197), score: 0.44 DPP8dipeptidyl-peptidase 8 (ENSG00000074603), score: 0.45 DPY19L3dpy-19-like 3 (C. elegans) (ENSG00000178904), score: 0.44 DRD2dopamine receptor D2 (ENSG00000149295), score: 0.56 DSCAMDown syndrome cell adhesion molecule (ENSG00000171587), score: 0.5 DUS3Ldihydrouridine synthase 3-like (S. cerevisiae) (ENSG00000141994), score: 0.53 ECHDC3enoyl CoA hydratase domain containing 3 (ENSG00000134463), score: -0.48 EIF4EBP1eukaryotic translation initiation factor 4E binding protein 1 (ENSG00000187840), score: -0.42 ELP2elongation protein 2 homolog (S. cerevisiae) (ENSG00000134759), score: 0.51 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENSG00000136960), score: 0.43 EPHA8EPH receptor A8 (ENSG00000070886), score: 0.5 EPHX4epoxide hydrolase 4 (ENSG00000172031), score: 0.45 ERAL1Era G-protein-like 1 (E. coli) (ENSG00000132591), score: -0.43 ERBB2v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) (ENSG00000141736), score: -0.44 EXD2exonuclease 3'-5' domain containing 2 (ENSG00000081177), score: 0.55 EXOSC1exosome component 1 (ENSG00000171311), score: -0.42 EXTL2exostoses (multiple)-like 2 (ENSG00000162694), score: 0.46 FADDFas (TNFRSF6)-associated via death domain (ENSG00000168040), score: -0.52 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: -0.53 FAM98Afamily with sequence similarity 98, member A (ENSG00000119812), score: -0.42 FBXO33F-box protein 33 (ENSG00000165355), score: 0.42 FBXO7F-box protein 7 (ENSG00000100225), score: -0.44 FGL2fibrinogen-like 2 (ENSG00000127951), score: -0.43 FHL3four and a half LIM domains 3 (ENSG00000183386), score: -0.48 FIBCD1fibrinogen C domain containing 1 (ENSG00000130720), score: 0.49 FRS2fibroblast growth factor receptor substrate 2 (ENSG00000166225), score: 0.52 GAAglucosidase, alpha; acid (ENSG00000171298), score: 0.43 GABRA1gamma-aminobutyric acid (GABA) A receptor, alpha 1 (ENSG00000022355), score: 0.43 GAD2glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) (ENSG00000136750), score: 0.47 GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: 0.59 GLRA1glycine receptor, alpha 1 (ENSG00000145888), score: 0.79 GLRBglycine receptor, beta (ENSG00000109738), score: 0.45 GNALguanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type (ENSG00000141404), score: 0.42 GNG13guanine nucleotide binding protein (G protein), gamma 13 (ENSG00000127588), score: 0.51 GPBP1L1GC-rich promoter binding protein 1-like 1 (ENSG00000159592), score: -0.51 GPC5glypican 5 (ENSG00000179399), score: 0.56 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.47 GRIA1glutamate receptor, ionotropic, AMPA 1 (ENSG00000155511), score: 0.45 GRIK4glutamate receptor, ionotropic, kainate 4 (ENSG00000149403), score: 0.45 GRIN2Bglutamate receptor, ionotropic, N-methyl D-aspartate 2B (ENSG00000150086), score: 0.46 GSG1LGSG1-like (ENSG00000169181), score: 0.48 HDAC11histone deacetylase 11 (ENSG00000163517), score: 0.42 HDHD3haloacid dehalogenase-like hydrolase domain containing 3 (ENSG00000119431), score: -0.43 HMBOX1homeobox containing 1 (ENSG00000147421), score: 0.59 HMG20Ahigh-mobility group 20A (ENSG00000140382), score: 0.45 HMHA1histocompatibility (minor) HA-1 (ENSG00000180448), score: -0.47 IGDCC4immunoglobulin superfamily, DCC subclass, member 4 (ENSG00000103742), score: 0.46 IGFBP7insulin-like growth factor binding protein 7 (ENSG00000163453), score: -0.48 IL12RB2interleukin 12 receptor, beta 2 (ENSG00000081985), score: 0.57 IL22interleukin 22 (ENSG00000127318), score: 0.5 ITPRIPinositol 1,4,5-triphosphate receptor interacting protein (ENSG00000148841), score: -0.42 IWS1IWS1 homolog (S. cerevisiae) (ENSG00000163166), score: -0.53 KCNA1potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) (ENSG00000111262), score: 0.46 KCNA6potassium voltage-gated channel, shaker-related subfamily, member 6 (ENSG00000151079), score: 0.55 KCNH7potassium voltage-gated channel, subfamily H (eag-related), member 7 (ENSG00000184611), score: 0.66 KCNJ13potassium inwardly-rectifying channel, subfamily J, member 13 (ENSG00000115474), score: 0.66 KCNK2potassium channel, subfamily K, member 2 (ENSG00000082482), score: 0.66 KCNK9potassium channel, subfamily K, member 9 (ENSG00000169427), score: 0.44 KIAA0240KIAA0240 (ENSG00000112624), score: 0.52 KIAA0391KIAA0391 (ENSG00000100890), score: -0.57 KIAA1279KIAA1279 (ENSG00000198954), score: 0.44 KIAA1467KIAA1467 (ENSG00000084444), score: 0.44 KLHL26kelch-like 26 (Drosophila) (ENSG00000167487), score: 0.43 KRT9keratin 9 (ENSG00000171403), score: 1 LCTlactase (ENSG00000115850), score: 0.44 LEPRE1leucine proline-enriched proteoglycan (leprecan) 1 (ENSG00000117385), score: -0.49 LGALS3lectin, galactoside-binding, soluble, 3 (ENSG00000131981), score: -0.58 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.51 LINGO2leucine rich repeat and Ig domain containing 2 (ENSG00000174482), score: 0.42 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.72 LMF2lipase maturation factor 2 (ENSG00000100258), score: -0.57 LONRF2LON peptidase N-terminal domain and ring finger 2 (ENSG00000170500), score: 0.5 LPPR4lipid phosphate phosphatase-related protein type 4 (ENSG00000117600), score: 0.43 LRRN1leucine rich repeat neuronal 1 (ENSG00000175928), score: 0.44 LRRTM3leucine rich repeat transmembrane neuronal 3 (ENSG00000198739), score: 0.46 LSM10LSM10, U7 small nuclear RNA associated (ENSG00000181817), score: -0.48 LYPD6LY6/PLAUR domain containing 6 (ENSG00000187123), score: 0.44 MAD2L2MAD2 mitotic arrest deficient-like 2 (yeast) (ENSG00000116670), score: -0.45 MAP3K14mitogen-activated protein kinase kinase kinase 14 (ENSG00000006062), score: -0.42 MAP9microtubule-associated protein 9 (ENSG00000164114), score: 0.45 MAS1MAS1 oncogene (ENSG00000130368), score: 0.53 MATKmegakaryocyte-associated tyrosine kinase (ENSG00000007264), score: 0.47 MC4Rmelanocortin 4 receptor (ENSG00000166603), score: 0.46 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.44 MICALL2MICAL-like 2 (ENSG00000164877), score: -0.46 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.43 MRPL16mitochondrial ribosomal protein L16 (ENSG00000166902), score: -0.48 MRPL21mitochondrial ribosomal protein L21 (ENSG00000197345), score: -0.54 MRPL34mitochondrial ribosomal protein L34 (ENSG00000130312), score: -0.53 MRPS11mitochondrial ribosomal protein S11 (ENSG00000181991), score: -0.54 MTM1myotubularin 1 (ENSG00000171100), score: -0.65 MTMR14myotubularin related protein 14 (ENSG00000163719), score: -0.44 MTMR6myotubularin related protein 6 (ENSG00000139505), score: 0.5 MYO16myosin XVI (ENSG00000041515), score: 0.57 NADSYN1NAD synthetase 1 (ENSG00000172890), score: -0.51 NAIF1nuclear apoptosis inducing factor 1 (ENSG00000171169), score: -0.46 NCKAP1NCK-associated protein 1 (ENSG00000061676), score: 0.45 NDST2N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 (ENSG00000166507), score: 0.44 NDST4N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 (ENSG00000138653), score: 0.56 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.65 NFE2L2nuclear factor (erythroid-derived 2)-like 2 (ENSG00000116044), score: -0.62 NIT2nitrilase family, member 2 (ENSG00000114021), score: -0.42 NKD1naked cuticle homolog 1 (Drosophila) (ENSG00000140807), score: 0.5 NKIRAS2NFKB inhibitor interacting Ras-like 2 (ENSG00000168256), score: -0.47 NLGN1neuroligin 1 (ENSG00000169760), score: 0.44 NPEPL1aminopeptidase-like 1 (ENSG00000215440), score: -0.53 NPY2Rneuropeptide Y receptor Y2 (ENSG00000185149), score: 0.44 NR2C2nuclear receptor subfamily 2, group C, member 2 (ENSG00000177463), score: 0.43 NRSN1neurensin 1 (ENSG00000152954), score: 0.45 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.7 NT5DC35'-nucleotidase domain containing 3 (ENSG00000111696), score: 0.51 NTHL1nth endonuclease III-like 1 (E. coli) (ENSG00000065057), score: -0.46 NUBPLnucleotide binding protein-like (ENSG00000151413), score: -0.57 NUP37nucleoporin 37kDa (ENSG00000075188), score: -0.46 OCIAD1OCIA domain containing 1 (ENSG00000109180), score: 0.43 OGFOD12-oxoglutarate and iron-dependent oxygenase domain containing 1 (ENSG00000087263), score: 0.56 OPTNoptineurin (ENSG00000123240), score: -0.67 OSTF1osteoclast stimulating factor 1 (ENSG00000134996), score: -0.43 OSTM1osteopetrosis associated transmembrane protein 1 (ENSG00000081087), score: 0.44 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: -0.49 PAOXpolyamine oxidase (exo-N4-amino) (ENSG00000148832), score: -0.49 PARP16poly (ADP-ribose) polymerase family, member 16 (ENSG00000138617), score: -0.47 PCDH20protocadherin 20 (ENSG00000197991), score: 0.53 PDE10Aphosphodiesterase 10A (ENSG00000112541), score: 0.56 PEMTphosphatidylethanolamine N-methyltransferase (ENSG00000133027), score: -0.53 PENKproenkephalin (ENSG00000181195), score: 0.49 PEX10peroxisomal biogenesis factor 10 (ENSG00000157911), score: -0.44 PEX11Gperoxisomal biogenesis factor 11 gamma (ENSG00000104883), score: -0.46 PEX5Lperoxisomal biogenesis factor 5-like (ENSG00000114757), score: 0.51 PGAP3post-GPI attachment to proteins 3 (ENSG00000161395), score: -0.43 PLEKHN1pleckstrin homology domain containing, family N member 1 (ENSG00000187583), score: 0.73 PPCSphosphopantothenoylcysteine synthetase (ENSG00000127125), score: -0.61 PPWD1peptidylprolyl isomerase domain and WD repeat containing 1 (ENSG00000113593), score: -0.54 PQLC1PQ loop repeat containing 1 (ENSG00000122490), score: -0.55 PRICKLE2prickle homolog 2 (Drosophila) (ENSG00000163637), score: 0.47 PRKG2protein kinase, cGMP-dependent, type II (ENSG00000138669), score: 0.49 PRMT8protein arginine methyltransferase 8 (ENSG00000111218), score: 0.45 PROX2prospero homeobox 2 (ENSG00000119608), score: 0.66 PSMG3proteasome (prosome, macropain) assembly chaperone 3 (ENSG00000157778), score: -0.42 PTPLAD2protein tyrosine phosphatase-like A domain containing 2 (ENSG00000188921), score: 0.53 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: -0.43 RAB20RAB20, member RAS oncogene family (ENSG00000139832), score: -0.45 RAB3GAP2RAB3 GTPase activating protein subunit 2 (non-catalytic) (ENSG00000118873), score: 0.43 RALGAPA1Ral GTPase activating protein, alpha subunit 1 (catalytic) (ENSG00000174373), score: 0.44 RASGEF1ARasGEF domain family, member 1A (ENSG00000198915), score: 0.42 RBM19RNA binding motif protein 19 (ENSG00000122965), score: -0.49 RGNregucalcin (senescence marker protein-30) (ENSG00000130988), score: -0.51 RGS20regulator of G-protein signaling 20 (ENSG00000147509), score: 0.53 RGS7BPregulator of G-protein signaling 7 binding protein (ENSG00000186479), score: 0.44 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.55 RGS9regulator of G-protein signaling 9 (ENSG00000108370), score: 0.6 RHBDF2rhomboid 5 homolog 2 (Drosophila) (ENSG00000129667), score: -0.44 RICTORRPTOR independent companion of MTOR, complex 2 (ENSG00000164327), score: 0.5 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (ENSG00000137275), score: -0.47 RIT2Ras-like without CAAX 2 (ENSG00000152214), score: 0.5 RPF1ribosome production factor 1 homolog (S. cerevisiae) (ENSG00000117133), score: -0.45 RPS19BP1ribosomal protein S19 binding protein 1 (ENSG00000187051), score: -0.42 RPUSD4RNA pseudouridylate synthase domain containing 4 (ENSG00000165526), score: -0.47 SCAMP1secretory carrier membrane protein 1 (ENSG00000085365), score: 0.49 SCAMP2secretory carrier membrane protein 2 (ENSG00000140497), score: -0.43 SCG2secretogranin II (ENSG00000171951), score: 0.44 SCRN2secernin 2 (ENSG00000141295), score: -0.48 SEC31BSEC31 homolog B (S. cerevisiae) (ENSG00000075826), score: -0.81 SEMA3Asema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A (ENSG00000075213), score: 0.5 SEMA3Esema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E (ENSG00000170381), score: 0.51 SESN3sestrin 3 (ENSG00000149212), score: 0.44 SETDB2SET domain, bifurcated 2 (ENSG00000136169), score: -0.6 SGK196protein kinase-like protein SgK196 (ENSG00000185900), score: 0.5 SGSM1small G protein signaling modulator 1 (ENSG00000167037), score: 0.43 SIRT5sirtuin 5 (ENSG00000124523), score: -0.42 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.51 SLC24A2solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 (ENSG00000155886), score: 0.47 SLC24A3solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 (ENSG00000185052), score: 0.46 SLC24A4solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 (ENSG00000140090), score: 0.52 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: -0.45 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.47 SLC36A4solute carrier family 36 (proton/amino acid symporter), member 4 (ENSG00000180773), score: 0.57 SLC39A12solute carrier family 39 (zinc transporter), member 12 (ENSG00000148482), score: 0.58 SLC5A7solute carrier family 5 (choline transporter), member 7 (ENSG00000115665), score: 0.71 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (ENSG00000132164), score: 0.58 SLC6A5solute carrier family 6 (neurotransmitter transporter, glycine), member 5 (ENSG00000165970), score: 0.63 SLC7A10solute carrier family 7, (neutral amino acid transporter, y+ system) member 10 (ENSG00000130876), score: 0.5 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (ENSG00000151012), score: 0.52 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (ENSG00000100335), score: -0.42 SNX16sorting nexin 16 (ENSG00000104497), score: 0.54 SPRED1sprouty-related, EVH1 domain containing 1 (ENSG00000166068), score: 0.52 SPRED2sprouty-related, EVH1 domain containing 2 (ENSG00000198369), score: 0.45 SRGAP3SLIT-ROBO Rho GTPase activating protein 3 (ENSG00000196220), score: 0.47 SSBP1single-stranded DNA binding protein 1 (ENSG00000106028), score: -0.62 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.47 SSTsomatostatin (ENSG00000157005), score: 0.49 ST3GAL2ST3 beta-galactoside alpha-2,3-sialyltransferase 2 (ENSG00000157350), score: 0.44 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (ENSG00000124214), score: -0.46 SV2Csynaptic vesicle glycoprotein 2C (ENSG00000122012), score: 0.62 SYNJ1synaptojanin 1 (ENSG00000159082), score: 0.47 TAC1tachykinin, precursor 1 (ENSG00000006128), score: 0.45 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (ENSG00000172660), score: -0.44 TATDN1TatD DNase domain containing 1 (ENSG00000147687), score: -0.44 TBC1D30TBC1 domain family, member 30 (ENSG00000111490), score: 0.5 TIRAPtoll-interleukin 1 receptor (TIR) domain containing adaptor protein (ENSG00000150455), score: -0.48 TLN1talin 1 (ENSG00000137076), score: -0.49 TMEM170Atransmembrane protein 170A (ENSG00000166822), score: -0.47 TMEM53transmembrane protein 53 (ENSG00000126106), score: -0.53 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (ENSG00000185215), score: -0.48 TNFRSF1Atumor necrosis factor receptor superfamily, member 1A (ENSG00000067182), score: -0.43 TOP3Atopoisomerase (DNA) III alpha (ENSG00000177302), score: -0.56 TRAM2translocation associated membrane protein 2 (ENSG00000065308), score: -0.56 TRHRthyrotropin-releasing hormone receptor (ENSG00000174417), score: 0.88 TRIM35tripartite motif-containing 35 (ENSG00000104228), score: 0.49 TRPC3transient receptor potential cation channel, subfamily C, member 3 (ENSG00000138741), score: 0.48 TRPC5transient receptor potential cation channel, subfamily C, member 5 (ENSG00000072315), score: 0.48 TRRAPtransformation/transcription domain-associated protein (ENSG00000196367), score: 0.44 TSHZ2teashirt zinc finger homeobox 2 (ENSG00000182463), score: 0.44 TSPAN2tetraspanin 2 (ENSG00000134198), score: 0.68 TXNDC16thioredoxin domain containing 16 (ENSG00000087301), score: 0.67 UBE2Wubiquitin-conjugating enzyme E2W (putative) (ENSG00000104343), score: 0.44 UBLCP1ubiquitin-like domain containing CTD phosphatase 1 (ENSG00000164332), score: 0.49 UNC80unc-80 homolog (C. elegans) (ENSG00000144406), score: 0.45 VPS41vacuolar protein sorting 41 homolog (S. cerevisiae) (ENSG00000006715), score: 0.44 VRK2vaccinia related kinase 2 (ENSG00000028116), score: -0.5 WBP1WW domain binding protein 1 (ENSG00000115274), score: -0.48 WBP4WW domain binding protein 4 (formin binding protein 21) (ENSG00000120688), score: 0.43 WDR7WD repeat domain 7 (ENSG00000091157), score: 0.43 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.57 YAF2YY1 associated factor 2 (ENSG00000015153), score: 0.57 YIPF3Yip1 domain family, member 3 (ENSG00000137207), score: -0.54 YTHDF2YTH domain family, member 2 (ENSG00000198492), score: -0.42 ZBTB11zinc finger and BTB domain containing 11 (ENSG00000066422), score: 0.48 ZBTB44zinc finger and BTB domain containing 44 (ENSG00000196323), score: -0.43 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.49 ZFYVE21zinc finger, FYVE domain containing 21 (ENSG00000100711), score: -0.53
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_cb_f_ca1 | mmu | cb | f | _ |
| mmu_cb_m1_ca1 | mmu | cb | m | 1 |
| mmu_br_m2_ca1 | mmu | br | m | 2 |
| mmu_br_f_ca1 | mmu | br | f | _ |
| mmu_br_m1_ca1 | mmu | br | m | 1 |