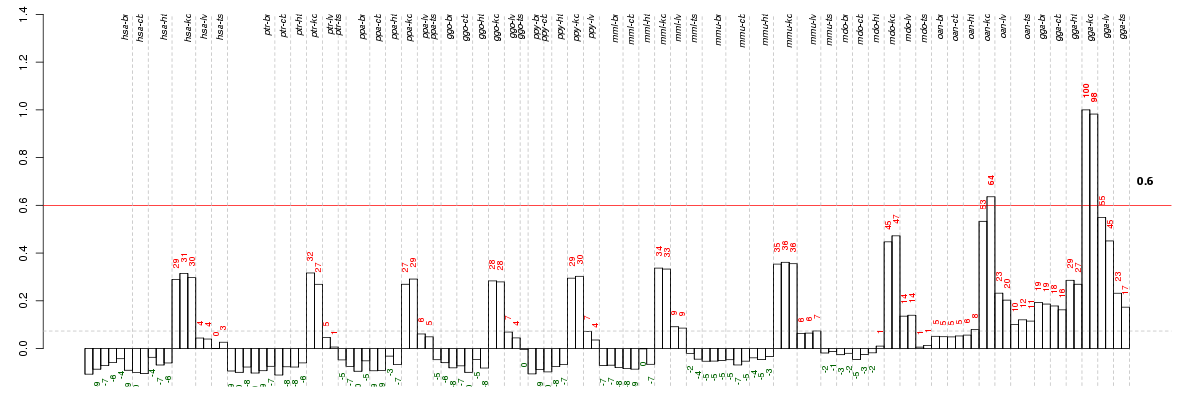

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
secretion
The controlled release of a substance by a cell, a group of cells, or a tissue.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
NA
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
sodium channel complex
An ion channel complex through which sodium ions pass.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
apical part of cell
The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
secondary active transmembrane transporter activity
Catalysis of the transfer of a solute from one side of a membrane to the other, up its concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction and is driven by a chemiosmotic source of energy. Chemiosmotic sources of energy include uniport, symport or antiport.
active transmembrane transporter activity
Catalysis of the transfer of a specific substance or related group of substances from one side of a membrane to the other, up the solute's concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04960 | 1.285e-02 | 0.9843 | 6 | 16 | Aldosterone-regulated sodium reabsorption |
ACAD8acyl-CoA dehydrogenase family, member 8 (ENSG00000151498), score: 0.44 ACSM3acyl-CoA synthetase medium-chain family member 3 (ENSG00000005187), score: 0.47 ACVR1activin A receptor, type I (ENSG00000115170), score: 0.38 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.39 ADAadenosine deaminase (ENSG00000196839), score: 0.41 ADAT2adenosine deaminase, tRNA-specific 2, TAD2 homolog (S. cerevisiae) (ENSG00000189007), score: 0.41 AGR2anterior gradient homolog 2 (Xenopus laevis) (ENSG00000106541), score: 0.6 AKAP12A kinase (PRKA) anchor protein 12 (ENSG00000131016), score: -0.32 AKAP6A kinase (PRKA) anchor protein 6 (ENSG00000151320), score: -0.3 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.37 ALKanaplastic lymphoma receptor tyrosine kinase (ENSG00000171094), score: 0.41 ANKS4Bankyrin repeat and sterile alpha motif domain containing 4B (ENSG00000175311), score: 0.42 AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.4 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: 0.36 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (ENSG00000113108), score: -0.48 APLP2amyloid beta (A4) precursor-like protein 2 (ENSG00000084234), score: 0.39 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (ENSG00000149182), score: -0.3 ARHGAP24Rho GTPase activating protein 24 (ENSG00000138639), score: 0.36 ARHGAP6Rho GTPase activating protein 6 (ENSG00000047648), score: 0.34 ARHGEF7Rho guanine nucleotide exchange factor (GEF) 7 (ENSG00000102606), score: -0.39 ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: -0.47 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ENSG00000153317), score: -0.31 ASXL2additional sex combs like 2 (Drosophila) (ENSG00000143970), score: 0.38 ATP6V0A4ATPase, H+ transporting, lysosomal V0 subunit a4 (ENSG00000105929), score: 0.43 ATP6V0D2ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 (ENSG00000147614), score: 0.34 ATP7AATPase, Cu++ transporting, alpha polypeptide (ENSG00000165240), score: 0.41 ATRNattractin (ENSG00000088812), score: 0.32 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.34 BAMBIBMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (ENSG00000095739), score: 0.35 BCAT1branched chain amino-acid transaminase 1, cytosolic (ENSG00000060982), score: 0.48 BMP3bone morphogenetic protein 3 (ENSG00000152785), score: 0.6 BPGM2,3-bisphosphoglycerate mutase (ENSG00000172331), score: -0.32 BRWD3bromodomain and WD repeat domain containing 3 (ENSG00000165288), score: 0.35 BSDC1BSD domain containing 1 (ENSG00000160058), score: -0.31 C13orf39chromosome 13 open reading frame 39 (ENSG00000139780), score: 0.34 C16orf7chromosome 16 open reading frame 7 (ENSG00000075399), score: -0.33 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.38 C20orf79chromosome 20 open reading frame 79 (ENSG00000132631), score: 0.34 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.47 C6orf186chromosome 6 open reading frame 186 (ENSG00000053328), score: 0.45 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.48 CALML4calmodulin-like 4 (ENSG00000129007), score: 0.41 CAPSLcalcyphosine-like (ENSG00000152611), score: 0.4 CASRcalcium-sensing receptor (ENSG00000036828), score: 0.33 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: 0.35 CCDC45coiled-coil domain containing 45 (ENSG00000141325), score: -0.33 CCNYcyclin Y (ENSG00000108100), score: -0.3 CD40LGCD40 ligand (ENSG00000102245), score: 0.72 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.36 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 0.57 CDH1cadherin 1, type 1, E-cadherin (epithelial) (ENSG00000039068), score: 0.34 CDK15cyclin-dependent kinase 15 (ENSG00000138395), score: 0.52 CDK6cyclin-dependent kinase 6 (ENSG00000105810), score: 0.42 CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: -0.37 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.41 CHST11carbohydrate (chondroitin 4) sulfotransferase 11 (ENSG00000171310), score: 0.36 CHST9carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 (ENSG00000154080), score: 0.42 CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.37 CINPcyclin-dependent kinase 2 interacting protein (ENSG00000100865), score: -0.39 CLCN4chloride channel 4 (ENSG00000073464), score: -0.34 CLDN16claudin 16 (ENSG00000113946), score: 0.41 CLDN19claudin 19 (ENSG00000164007), score: 0.33 CLIC6chloride intracellular channel 6 (ENSG00000159212), score: 0.66 CLPXClpX caseinolytic peptidase X homolog (E. coli) (ENSG00000166855), score: 0.38 CLUclusterin (ENSG00000120885), score: -0.33 CNNM2cyclin M2 (ENSG00000148842), score: 0.5 COL4A4collagen, type IV, alpha 4 (ENSG00000081052), score: 0.32 CPPED1calcineurin-like phosphoesterase domain containing 1 (ENSG00000103381), score: 0.37 CRBNcereblon (ENSG00000113851), score: -0.3 CRISP1cysteine-rich secretory protein 1 (ENSG00000124812), score: 0.96 CTDSPLCTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like (ENSG00000144677), score: 0.39 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.38 CUBNcubilin (intrinsic factor-cobalamin receptor) (ENSG00000107611), score: 0.48 CUEDC2CUE domain containing 2 (ENSG00000107874), score: -0.32 DAB2disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) (ENSG00000153071), score: 0.32 DACT2dapper, antagonist of beta-catenin, homolog 2 (Xenopus laevis) (ENSG00000164488), score: 0.36 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.42 DCAF17DDB1 and CUL4 associated factor 17 (ENSG00000115827), score: -0.34 DCUN1D5DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) (ENSG00000137692), score: -0.46 DDX1DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (ENSG00000079785), score: -0.31 DDX24DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 (ENSG00000089737), score: -0.34 DENND2DDENN/MADD domain containing 2D (ENSG00000162777), score: 0.36 DENND5ADENN/MADD domain containing 5A (ENSG00000184014), score: -0.37 DHDPSLdihydrodipicolinate synthase-like, mitochondrial (ENSG00000241935), score: 0.35 DHX29DEAH (Asp-Glu-Ala-His) box polypeptide 29 (ENSG00000067248), score: -0.32 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: -0.35 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (ENSG00000168259), score: -0.32 DNMBPdynamin binding protein (ENSG00000107554), score: 0.35 DPP4dipeptidyl-peptidase 4 (ENSG00000197635), score: 0.34 DRAM2DNA-damage regulated autophagy modulator 2 (ENSG00000156171), score: 0.38 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.32 EHFets homologous factor (ENSG00000135373), score: 0.45 ELP4elongation protein 4 homolog (S. cerevisiae) (ENSG00000109911), score: -0.31 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENSG00000136960), score: -0.32 EPB41L4Aerythrocyte membrane protein band 4.1 like 4A (ENSG00000129595), score: 0.33 EPCAMepithelial cell adhesion molecule (ENSG00000119888), score: 0.35 EPS15L1epidermal growth factor receptor pathway substrate 15-like 1 (ENSG00000127527), score: -0.36 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (ENSG00000178568), score: 0.45 EREGepiregulin (ENSG00000124882), score: 0.92 ERI1exoribonuclease 1 (ENSG00000104626), score: 0.39 ESR2estrogen receptor 2 (ER beta) (ENSG00000140009), score: 0.53 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.39 FAM105Afamily with sequence similarity 105, member A (ENSG00000145569), score: 0.41 FAM109Bfamily with sequence similarity 109, member B (ENSG00000177096), score: 0.41 FAM166Afamily with sequence similarity 166, member A (ENSG00000188163), score: -0.31 FAM3Dfamily with sequence similarity 3, member D (ENSG00000198643), score: 0.42 FAM82A1family with sequence similarity 82, member A1 (ENSG00000115841), score: 0.34 FANCAFanconi anemia, complementation group A (ENSG00000187741), score: -0.36 FBP2fructose-1,6-bisphosphatase 2 (ENSG00000130957), score: 0.4 FBXL18F-box and leucine-rich repeat protein 18 (ENSG00000155034), score: 0.39 FBXL20F-box and leucine-rich repeat protein 20 (ENSG00000108306), score: 0.44 FER1L5fer-1-like 5 (C. elegans) (ENSG00000214272), score: 0.4 FER1L6fer-1-like 6 (C. elegans) (ENSG00000214814), score: 0.33 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.44 FEZ2fasciculation and elongation protein zeta 2 (zygin II) (ENSG00000171055), score: -0.32 FGF22fibroblast growth factor 22 (ENSG00000070388), score: 0.36 FNDC4fibronectin type III domain containing 4 (ENSG00000115226), score: -0.32 FOXI1forkhead box I1 (ENSG00000168269), score: 0.37 FRAS1Fraser syndrome 1 (ENSG00000138759), score: 0.46 FSHBfollicle stimulating hormone, beta polypeptide (ENSG00000131808), score: 0.37 FTSJD2FtsJ methyltransferase domain containing 2 (ENSG00000137200), score: -0.33 FYNFYN oncogene related to SRC, FGR, YES (ENSG00000010810), score: -0.34 GALNT14UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) (ENSG00000158089), score: 0.36 GALNT5UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) (ENSG00000136542), score: 0.46 GATA3GATA binding protein 3 (ENSG00000107485), score: 0.48 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.32 GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: -0.35 GPR149G protein-coupled receptor 149 (ENSG00000174948), score: 0.55 GPR64G protein-coupled receptor 64 (ENSG00000173698), score: 0.42 GRHL2grainyhead-like 2 (Drosophila) (ENSG00000083307), score: 0.5 GRK5G protein-coupled receptor kinase 5 (ENSG00000198873), score: -0.41 GSCgoosecoid homeobox (ENSG00000133937), score: 0.72 GTPBP2GTP binding protein 2 (ENSG00000172432), score: -0.31 HEATR6HEAT repeat containing 6 (ENSG00000068097), score: 0.34 HMCN1hemicentin 1 (ENSG00000143341), score: 0.45 HNF4Ghepatocyte nuclear factor 4, gamma (ENSG00000164749), score: 0.56 HOXA10homeobox A10 (ENSG00000153807), score: 0.41 HOXA13homeobox A13 (ENSG00000106031), score: 0.77 HOXB7homeobox B7 (ENSG00000120087), score: 0.37 HOXD10homeobox D10 (ENSG00000128710), score: 0.66 HOXD4homeobox D4 (ENSG00000170166), score: 0.44 HPGDShematopoietic prostaglandin D synthase (ENSG00000163106), score: 0.71 HSD11B2hydroxysteroid (11-beta) dehydrogenase 2 (ENSG00000176387), score: 0.41 HSF4heat shock transcription factor 4 (ENSG00000102878), score: -0.33 ICOSinducible T-cell co-stimulator (ENSG00000163600), score: 0.63 IKZF3IKAROS family zinc finger 3 (Aiolos) (ENSG00000161405), score: 0.45 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.34 IL7Rinterleukin 7 receptor (ENSG00000168685), score: 0.33 IMPG2interphotoreceptor matrix proteoglycan 2 (ENSG00000081148), score: 0.4 INTS2integrator complex subunit 2 (ENSG00000108506), score: 0.34 IRF8interferon regulatory factor 8 (ENSG00000140968), score: 0.33 JAG1jagged 1 (ENSG00000101384), score: 0.43 JOSD1Josephin domain containing 1 (ENSG00000100221), score: -0.32 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (ENSG00000171385), score: -0.3 KCNE2potassium voltage-gated channel, Isk-related family, member 2 (ENSG00000159197), score: 0.33 KCNJ1potassium inwardly-rectifying channel, subfamily J, member 1 (ENSG00000151704), score: 0.32 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (ENSG00000153822), score: 0.44 KIAA1522KIAA1522 (ENSG00000162522), score: 0.37 KIAA1737KIAA1737 (ENSG00000198894), score: -0.34 KIAA1841KIAA1841 (ENSG00000162929), score: -0.31 KIRRELkin of IRRE like (Drosophila) (ENSG00000183853), score: 0.41 KLHDC1kelch domain containing 1 (ENSG00000197776), score: 0.32 KLHDC8Bkelch domain containing 8B (ENSG00000185909), score: -0.31 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.56 KLHL23kelch-like 23 (Drosophila) (ENSG00000213160), score: -0.31 KRT9keratin 9 (ENSG00000171403), score: 0.34 LACTBlactamase, beta (ENSG00000103642), score: 0.34 LAD1ladinin 1 (ENSG00000159166), score: 0.42 LAMA1laminin, alpha 1 (ENSG00000101680), score: 0.45 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.54 LASP1LIM and SH3 protein 1 (ENSG00000002834), score: 0.45 LASS6LAG1 homolog, ceramide synthase 6 (ENSG00000172292), score: -0.43 LCMT1leucine carboxyl methyltransferase 1 (ENSG00000205629), score: -0.34 LGMNlegumain (ENSG00000100600), score: 0.37 LMX1BLIM homeobox transcription factor 1, beta (ENSG00000136944), score: 0.41 LOC100134052similar to ubiquitin associated and SH3 domain containing, A (ENSG00000160185), score: 0.32 LRP11low density lipoprotein receptor-related protein 11 (ENSG00000120256), score: -0.36 LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.31 LRRC58leucine rich repeat containing 58 (ENSG00000163428), score: 0.53 LY75lymphocyte antigen 75 (ENSG00000054219), score: 0.32 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.36 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: 0.36 MAP3K2mitogen-activated protein kinase kinase kinase 2 (ENSG00000169967), score: 0.38 MARVELD2MARVEL domain containing 2 (ENSG00000152939), score: 0.32 MARVELD3MARVEL domain containing 3 (ENSG00000140832), score: 0.37 MBD3methyl-CpG binding domain protein 3 (ENSG00000071655), score: -0.37 MED13mediator complex subunit 13 (ENSG00000108510), score: 0.33 MFI2antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 (ENSG00000163975), score: 0.37 MOCS1molybdenum cofactor synthesis 1 (ENSG00000124615), score: 0.33 MPDZmultiple PDZ domain protein (ENSG00000107186), score: -0.31 MPZL3myelin protein zero-like 3 (ENSG00000160588), score: 0.37 MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.4 MTA1metastasis associated 1 (ENSG00000182979), score: -0.35 MYO1Dmyosin ID (ENSG00000176658), score: 0.38 NFAT5nuclear factor of activated T-cells 5, tonicity-responsive (ENSG00000102908), score: 0.35 NHSNance-Horan syndrome (congenital cataracts and dental anomalies) (ENSG00000188158), score: 0.37 NKIRAS2NFKB inhibitor interacting Ras-like 2 (ENSG00000168256), score: 0.32 NLRC3NLR family, CARD domain containing 3 (ENSG00000167984), score: 0.37 NOBOXNOBOX oogenesis homeobox (ENSG00000106410), score: 0.42 NPHS2nephrosis 2, idiopathic, steroid-resistant (podocin) (ENSG00000116218), score: 0.36 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (ENSG00000113389), score: 0.49 NRBP1nuclear receptor binding protein 1 (ENSG00000115216), score: -0.52 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: -0.37 NT5C35'-nucleotidase, cytosolic III (ENSG00000122643), score: 0.36 OLFML2Aolfactomedin-like 2A (ENSG00000185585), score: 0.35 ORC2Lorigin recognition complex, subunit 2-like (yeast) (ENSG00000115942), score: -0.35 ORC5Lorigin recognition complex, subunit 5-like (yeast) (ENSG00000164815), score: -0.3 OSTalphaorganic solute transporter alpha (ENSG00000163959), score: 0.33 PAIP1poly(A) binding protein interacting protein 1 (ENSG00000172239), score: -0.42 PANX3pannexin 3 (ENSG00000154143), score: 0.38 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.4 PAQR5progestin and adipoQ receptor family member V (ENSG00000137819), score: 0.39 PARVBparvin, beta (ENSG00000188677), score: -0.36 PATZ1POZ (BTB) and AT hook containing zinc finger 1 (ENSG00000100105), score: -0.41 PAX9paired box 9 (ENSG00000198807), score: 0.33 PDCD5programmed cell death 5 (ENSG00000105185), score: -0.41 PDE4Bphosphodiesterase 4B, cAMP-specific (ENSG00000184588), score: -0.37 PDX1pancreatic and duodenal homeobox 1 (ENSG00000139515), score: 0.79 PEPDpeptidase D (ENSG00000124299), score: 0.35 PHEXphosphate regulating endopeptidase homolog, X-linked (ENSG00000102174), score: 0.7 PHF16PHD finger protein 16 (ENSG00000102221), score: 0.33 PHKA2phosphorylase kinase, alpha 2 (liver) (ENSG00000044446), score: -0.32 PIK3CGphosphoinositide-3-kinase, catalytic, gamma polypeptide (ENSG00000105851), score: 0.44 PLA2G15phospholipase A2, group XV (ENSG00000103066), score: 0.38 PLEKHF2pleckstrin homology domain containing, family F (with FYVE domain) member 2 (ENSG00000175895), score: 0.33 PLEKHH2pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 (ENSG00000152527), score: 0.36 PLRG1pleiotropic regulator 1 (PRL1 homolog, Arabidopsis) (ENSG00000171566), score: -0.46 PM20D2peptidase M20 domain containing 2 (ENSG00000146281), score: 0.42 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (ENSG00000100479), score: 0.38 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (ENSG00000125630), score: 0.35 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta (ENSG00000175470), score: -0.33 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta (ENSG00000112640), score: -0.36 PRKAG3protein kinase, AMP-activated, gamma 3 non-catalytic subunit (ENSG00000115592), score: 0.32 PTDSS1phosphatidylserine synthase 1 (ENSG00000156471), score: -0.33 PTERphosphotriesterase related (ENSG00000165983), score: 0.39 PTPN7protein tyrosine phosphatase, non-receptor type 7 (ENSG00000143851), score: 0.32 PTPRQprotein tyrosine phosphatase, receptor type, Q (ENSG00000139304), score: 0.58 PYROXD2pyridine nucleotide-disulphide oxidoreductase domain 2 (ENSG00000119943), score: 0.46 RAB19RAB19, member RAS oncogene family (ENSG00000146955), score: 0.66 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.37 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.58 RBKSribokinase (ENSG00000171174), score: 0.38 RGNregucalcin (senescence marker protein-30) (ENSG00000130988), score: 0.47 RHAGRh-associated glycoprotein (ENSG00000112077), score: 0.78 RHCGRh family, C glycoprotein (ENSG00000140519), score: 0.44 RNASELribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) (ENSG00000135828), score: 0.35 ROR1receptor tyrosine kinase-like orphan receptor 1 (ENSG00000185483), score: 0.34 RPIAribose 5-phosphate isomerase A (ENSG00000153574), score: 0.4 RPS6KA2ribosomal protein S6 kinase, 90kDa, polypeptide 2 (ENSG00000071242), score: -0.35 RRM1ribonucleotide reductase M1 (ENSG00000167325), score: -0.46 RUSC2RUN and SH3 domain containing 2 (ENSG00000198853), score: -0.41 RXFP3relaxin/insulin-like family peptide receptor 3 (ENSG00000182631), score: 1 SALL1sal-like 1 (Drosophila) (ENSG00000103449), score: 0.38 SASH3SAM and SH3 domain containing 3 (ENSG00000122122), score: 0.36 SCINscinderin (ENSG00000006747), score: 0.34 SCNN1Asodium channel, nonvoltage-gated 1 alpha (ENSG00000111319), score: 0.33 SCNN1Bsodium channel, nonvoltage-gated 1, beta (ENSG00000168447), score: 0.47 SCNN1Gsodium channel, nonvoltage-gated 1, gamma (ENSG00000166828), score: 0.37 SDCCAG8serologically defined colon cancer antigen 8 (ENSG00000054282), score: -0.4 SDSLserine dehydratase-like (ENSG00000139410), score: 0.34 SEMA3Dsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D (ENSG00000153993), score: 0.37 SEMA5Asema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A (ENSG00000112902), score: 0.41 SEPT2septin 2 (ENSG00000168385), score: -0.31 SESN1sestrin 1 (ENSG00000080546), score: 0.34 SH2D4ASH2 domain containing 4A (ENSG00000104611), score: 0.35 SHISA2shisa homolog 2 (Xenopus laevis) (ENSG00000180730), score: 0.57 SIM1single-minded homolog 1 (Drosophila) (ENSG00000112246), score: 0.68 SIM2single-minded homolog 2 (Drosophila) (ENSG00000159263), score: 0.45 SLBPstem-loop binding protein (ENSG00000163950), score: -0.36 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.37 SLC13A3solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 (ENSG00000158296), score: 0.34 SLC16A12solute carrier family 16, member 12 (monocarboxylic acid transporter 12) (ENSG00000152779), score: 0.39 SLC16A2solute carrier family 16, member 2 (monocarboxylic acid transporter 8) (ENSG00000147100), score: 0.35 SLC16A4solute carrier family 16, member 4 (monocarboxylic acid transporter 5) (ENSG00000168679), score: 0.46 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.36 SLC20A1solute carrier family 20 (phosphate transporter), member 1 (ENSG00000144136), score: 0.41 SLC24A6solute carrier family 24 (sodium/potassium/calcium exchanger), member 6 (ENSG00000089060), score: 0.38 SLC25A21solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 (ENSG00000183032), score: 0.36 SLC25A48solute carrier family 25, member 48 (ENSG00000145832), score: 0.47 SLC34A1solute carrier family 34 (sodium phosphate), member 1 (ENSG00000131183), score: 0.49 SLC35F2solute carrier family 35, member F2 (ENSG00000110660), score: 0.35 SLC36A4solute carrier family 36 (proton/amino acid symporter), member 4 (ENSG00000180773), score: -0.33 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (ENSG00000080493), score: 0.32 SLC5A11solute carrier family 5 (sodium/glucose cotransporter), member 11 (ENSG00000158865), score: 0.37 SLC5A9solute carrier family 5 (sodium/glucose cotransporter), member 9 (ENSG00000117834), score: 0.33 SLC6A19solute carrier family 6 (neutral amino acid transporter), member 19 (ENSG00000174358), score: 0.51 SMAD6SMAD family member 6 (ENSG00000137834), score: 0.37 SMARCA5SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 (ENSG00000153147), score: -0.32 SMYD2SET and MYND domain containing 2 (ENSG00000143499), score: -0.31 SNTB2syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) (ENSG00000168807), score: 0.33 SNX24sorting nexin 24 (ENSG00000064652), score: 0.38 SNX33sorting nexin 33 (ENSG00000173548), score: 0.32 SOS2son of sevenless homolog 2 (Drosophila) (ENSG00000100485), score: 0.33 SPDEFSAM pointed domain containing ets transcription factor (ENSG00000124664), score: 0.36 SPHK1sphingosine kinase 1 (ENSG00000176170), score: 0.33 SPPL2Bsignal peptide peptidase-like 2B (ENSG00000005206), score: -0.37 SRMSsrc-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites (ENSG00000125508), score: 0.72 SRSF4serine/arginine-rich splicing factor 4 (ENSG00000116350), score: -0.33 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (ENSG00000184402), score: -0.31 ST14suppression of tumorigenicity 14 (colon carcinoma) (ENSG00000149418), score: 0.45 ST6GALNAC6ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 (ENSG00000160408), score: -0.31 STIM2stromal interaction molecule 2 (ENSG00000109689), score: -0.32 STK10serine/threonine kinase 10 (ENSG00000072786), score: 0.33 STK24serine/threonine kinase 24 (ENSG00000102572), score: -0.34 SUSD2sushi domain containing 2 (ENSG00000099994), score: 0.38 SUSD3sushi domain containing 3 (ENSG00000157303), score: 0.38 SYDE2synapse defective 1, Rho GTPase, homolog 2 (C. elegans) (ENSG00000097096), score: 0.34 SYPL1synaptophysin-like 1 (ENSG00000008282), score: -0.37 TADA2Atranscriptional adaptor 2A (ENSG00000108264), score: -0.32 TAF3TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa (ENSG00000165632), score: 0.34 TDRKHtudor and KH domain containing (ENSG00000182134), score: -0.31 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.41 TFAP2Btranscription factor AP-2 beta (activating enhancer binding protein 2 beta) (ENSG00000008196), score: 0.48 TFCP2L1transcription factor CP2-like 1 (ENSG00000115112), score: 0.35 THNSL2threonine synthase-like 2 (S. cerevisiae) (ENSG00000144115), score: 0.38 TINAGtubulointerstitial nephritis antigen (ENSG00000137251), score: 0.41 TM7SF4transmembrane 7 superfamily member 4 (ENSG00000164935), score: 0.35 TMCO3transmembrane and coiled-coil domains 3 (ENSG00000150403), score: -0.31 TMED8transmembrane emp24 protein transport domain containing 8 (ENSG00000100580), score: 0.59 TMEM120Btransmembrane protein 120B (ENSG00000188735), score: -0.34 TMEM171transmembrane protein 171 (ENSG00000157111), score: 0.48 TMEM199transmembrane protein 199 (ENSG00000244045), score: 0.37 TMEM27transmembrane protein 27 (ENSG00000147003), score: 0.39 TMEM51transmembrane protein 51 (ENSG00000171729), score: 0.35 TMIGD1transmembrane and immunoglobulin domain containing 1 (ENSG00000182271), score: 0.32 TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.58 TMPRSS2transmembrane protease, serine 2 (ENSG00000184012), score: 0.35 TMPRSS4transmembrane protease, serine 4 (ENSG00000137648), score: 0.32 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: -0.34 TNFRSF11Atumor necrosis factor receptor superfamily, member 11a, NFKB activator (ENSG00000141655), score: 0.41 TRAT1T cell receptor associated transmembrane adaptor 1 (ENSG00000163519), score: 0.49 TRPM6transient receptor potential cation channel, subfamily M, member 6 (ENSG00000119121), score: 0.37 TSG101tumor susceptibility gene 101 (ENSG00000074319), score: 0.35 TTC1tetratricopeptide repeat domain 1 (ENSG00000113312), score: -0.43 TTLL1tubulin tyrosine ligase-like family, member 1 (ENSG00000100271), score: -0.32 UBA2ubiquitin-like modifier activating enzyme 2 (ENSG00000126261), score: -0.41 UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.4 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: -0.37 UGCGUDP-glucose ceramide glucosyltransferase (ENSG00000148154), score: -0.36 VPS16vacuolar protein sorting 16 homolog (S. cerevisiae) (ENSG00000215305), score: 0.32 WDFY4WDFY family member 4 (ENSG00000128815), score: 0.52 WDR41WD repeat domain 41 (ENSG00000164253), score: -0.53 WDR8WD repeat domain 8 (ENSG00000116213), score: -0.32 WISP1WNT1 inducible signaling pathway protein 1 (ENSG00000104415), score: 0.44 WNT5Awingless-type MMTV integration site family, member 5A (ENSG00000114251), score: 0.38 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.39 XPR1xenotropic and polytropic retrovirus receptor 1 (ENSG00000143324), score: 0.38 YPEL5yippee-like 5 (Drosophila) (ENSG00000119801), score: -0.38 YWHAHtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide (ENSG00000128245), score: -0.31 ZBTB33zinc finger and BTB domain containing 33 (ENSG00000177485), score: 0.37 ZBTB37zinc finger and BTB domain containing 37 (ENSG00000185278), score: 0.34 ZBTB8Azinc finger and BTB domain containing 8A (ENSG00000160062), score: 0.34 ZBTB8Bzinc finger and BTB domain containing 8B (ENSG00000215897), score: 0.45 ZEB1zinc finger E-box binding homeobox 1 (ENSG00000148516), score: -0.34 ZFAND5zinc finger, AN1-type domain 5 (ENSG00000107372), score: -0.44 ZFPM2zinc finger protein, multitype 2 (ENSG00000169946), score: -0.31 ZHX3zinc fingers and homeoboxes 3 (ENSG00000174306), score: 0.33 ZMYM2zinc finger, MYM-type 2 (ENSG00000121741), score: -0.34 ZNF704zinc finger protein 704 (ENSG00000164684), score: 0.63 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.31
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_kd_f_ca1 | oan | kd | f | _ |
| gga_kd_f_ca1 | gga | kd | f | _ |
| gga_kd_m_ca1 | gga | kd | m | _ |