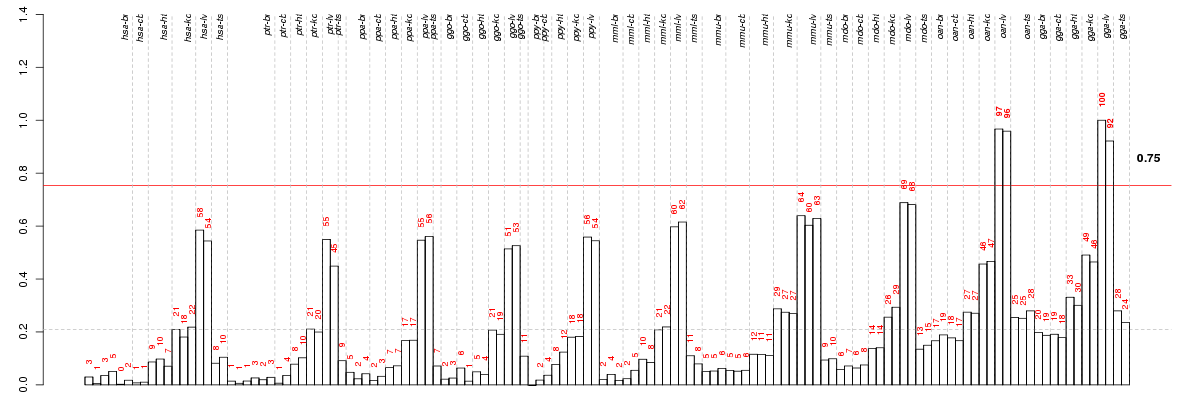

Under-expression is coded with green,
over-expression with red color.

transition metal ion transport
The directed movement of transition metal ions into, out of, within or between cells by means of some external agent such as a transporter or pore. A transition metal is an element whose atom has an incomplete d-subshell of extranuclear electrons, or which gives rise to a cation or cations with an incomplete d-subshell. Transition metals often have more than one valency state. Biologically relevant transition metals include vanadium, manganese, iron, copper, cobalt, nickel, molybdenum and silver.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
adaptive immune response
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process, and allowing for enhanced response to subsequent exposures to the same antigen (immunological memory).
immune effector process
Any process of the immune system that occurs as part of an immune response.
activation of immune response
Any process that initiates an immune response.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
leukocyte mediated immunity
Any process involved in the carrying out of an immune response by a leukocyte.
lymphocyte mediated immunity
Any process involved in the carrying out of an immune response by a lymphocyte.
humoral immune response mediated by circulating immunoglobulin
An immune response dependent upon secreted immunoglobulin. An example of this process is found in Mus musculus.
adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process that includes somatic recombination of germline gene segments encoding immunoglobulin superfamily domains, and allowing for enhanced responses upon subsequent exposures to the same antigen (immunological memory). Recombined receptors for antigen encoded by immunoglobulin superfamily domains include T cell receptors and immunoglobulins (antibodies). An example of this is the adaptive immune response found in Mus musculus.
acute inflammatory response
Inflammation which comprises a rapid, short-lived, relatively uniform response to acute injury or antigenic challenge and is characterized by accumulations of fluid, plasma proteins, and granulocytic leukocytes. An acute inflammatory response occurs within a matter of minutes or hours, and either resolves within a few days or becomes a chronic inflammatory response.
activation of plasma proteins involved in acute inflammatory response
Any process activating plasma proteins by proteolysis as part of an acute inflammatory response.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
protein maturation by peptide bond cleavage
The hydrolysis of a peptide bond or bonds within a protein as part of protein maturation, the process leading to the attainment of the full functional capacity of a protein.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cellular amino acid derivative metabolic process
The chemical reactions and pathways involving compounds derived from amino acids, organic acids containing one or more amino substituents.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
cellular aromatic compound metabolic process
The chemical reactions and pathways involving aromatic compounds, any organic compound characterized by one or more planar rings, each of which contains conjugated double bonds and delocalized pi electrons, as carried out by individual cells.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cation transport
The directed movement of cations, atoms or small molecules with a net positive charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
metal ion transport
The directed movement of metal ions, any metal ion with an electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
zinc ion transport
The directed movement of zinc (Zn) ions into, out of, within or between cells by means of some external agent such as a transporter or pore.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
complement activation, alternative pathway
Any process involved in the activation of any of the steps of the alternative pathway of the complement cascade which allows for the direct killing of microbes and the regulation of other immune processes.
complement activation, classical pathway
Any process involved in the activation of any of the steps of the classical pathway of the complement cascade which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes.
humoral immune response
An immune response mediated through a body fluid.
hemostasis
The stopping of bleeding (loss of body fluid) or the arrest of the circulation to an organ or part.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
cellular amino acid catabolic process
The chemical reactions and pathways resulting in the breakdown of amino acids, organic acids containing one or more amino substituents.
serine family amino acid metabolic process
The chemical reactions and pathways involving amino acids of the serine family, comprising cysteine, glycine, homoserine, selenocysteine and serine.
aromatic amino acid family metabolic process
The chemical reactions and pathways involving aromatic amino acid family, amino acids with aromatic ring (phenylalanine, tyrosine, tryptophan).
aromatic amino acid family catabolic process
The chemical reactions and pathways resulting in the breakdown of aromatic amino acid family, amino acids with aromatic ring (phenylalanine, tyrosine, tryptophan).
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
amine catabolic process
The chemical reactions and pathways resulting in the breakdown of any organic compound that is weakly basic in character and contains an amino or a substituted amino group. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
immunoglobulin mediated immune response
An immune response mediated by immunoglobulins, whether cell-bound or in solution.
protein processing
Any protein maturation process achieved by the cleavage of peptide bonds.
aromatic compound catabolic process
The chemical reactions and pathways resulting in the breakdown of aromatic compounds, any substance containing an aromatic carbon ring.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
monocarboxylic acid metabolic process
The chemical reactions and pathways involving monocarboxylic acids, any organic acid containing one carboxyl (COOH) group or anion (COO-).
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
wound healing
The series of events that restore integrity to a damaged tissue, following an injury.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
coagulation
The process by which a fluid solution, or part of it, changes into a solid or semisolid mass.
regulation of body fluid levels
Any process that modulates the levels of body fluids.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
cofactor metabolic process
The chemical reactions and pathways involving a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
protein maturation
Any process leading to the attainment of the full functional capacity of a protein.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
regulation of response to stress
Any process that modulates the frequency, rate or extent of a response to stress. Response to stress is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
immune effector process
Any process of the immune system that occurs as part of an immune response.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of response to stress
Any process that modulates the frequency, rate or extent of a response to stress. Response to stress is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
regulation of body fluid levels
Any process that modulates the levels of body fluids.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
protein maturation
Any process leading to the attainment of the full functional capacity of a protein.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
aromatic compound catabolic process
The chemical reactions and pathways resulting in the breakdown of aromatic compounds, any substance containing an aromatic carbon ring.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
activation of immune response
Any process that initiates an immune response.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
complement activation, alternative pathway
Any process involved in the activation of any of the steps of the alternative pathway of the complement cascade which allows for the direct killing of microbes and the regulation of other immune processes.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cellular amino acid catabolic process
The chemical reactions and pathways resulting in the breakdown of amino acids, organic acids containing one or more amino substituents.
aromatic amino acid family metabolic process
The chemical reactions and pathways involving aromatic amino acid family, amino acids with aromatic ring (phenylalanine, tyrosine, tryptophan).
aromatic amino acid family catabolic process
The chemical reactions and pathways resulting in the breakdown of aromatic amino acid family, amino acids with aromatic ring (phenylalanine, tyrosine, tryptophan).
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
complement activation, classical pathway
Any process involved in the activation of any of the steps of the classical pathway of the complement cascade which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
aromatic amino acid family catabolic process
The chemical reactions and pathways resulting in the breakdown of aromatic amino acid family, amino acids with aromatic ring (phenylalanine, tyrosine, tryptophan).
cellular amino acid catabolic process
The chemical reactions and pathways resulting in the breakdown of amino acids, organic acids containing one or more amino substituents.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
activation of plasma proteins involved in acute inflammatory response
Any process activating plasma proteins by proteolysis as part of an acute inflammatory response.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
humoral immune response mediated by circulating immunoglobulin
An immune response dependent upon secreted immunoglobulin. An example of this process is found in Mus musculus.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
organelle membrane
The lipid bilayer surrounding an organelle.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
vitamin binding
Interacting selectively and non-covalently with a vitamin, one of a number of unrelated organic substances that occur in many foods in small amounts and that are necessary in trace amounts for the normal metabolic functioning of the body.
pyridoxal phosphate binding
Interacting selectively and non-covalently with pyridoxal 5' phosphate, 3-hydroxy-5-(hydroxymethyl)-2-methyl4-pyridine carboxaldehyde 5' phosphate, the biologically active form of vitamin B6.
cofactor binding
Interacting selectively and non-covalently with a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
vitamin B6 binding
Interacting selectively and non-covalently with any of the vitamin B6 compounds: pyridoxal, pyridoxamine and pyridoxine and the active form, pyridoxal phosphate.
all
NA
pyridoxal phosphate binding
Interacting selectively and non-covalently with pyridoxal 5' phosphate, 3-hydroxy-5-(hydroxymethyl)-2-methyl4-pyridine carboxaldehyde 5' phosphate, the biologically active form of vitamin B6.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04610 | 9.161e-08 | 2.001 | 14 | 21 | Complement and coagulation cascades |
| 04141 | 1.353e-02 | 5.622 | 15 | 59 | Protein processing in endoplasmic reticulum |
| 00120 | 3.322e-02 | 0.2859 | 3 | 3 | Primary bile acid biosynthesis |
| 01100 | 3.492e-02 | 33.64 | 50 | 353 | Metabolic pathways |
A1CFAPOBEC1 complementation factor (ENSG00000148584), score: 0.43 A2LD1AIG2-like domain 1 (ENSG00000134864), score: 0.44 ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (ENSG00000165029), score: 0.43 ABCA12ATP-binding cassette, sub-family A (ABC1), member 12 (ENSG00000144452), score: 0.63 ABCB11ATP-binding cassette, sub-family B (MDR/TAP), member 11 (ENSG00000073734), score: 0.48 ABCC2ATP-binding cassette, sub-family C (CFTR/MRP), member 2 (ENSG00000023839), score: 0.45 ABHD5abhydrolase domain containing 5 (ENSG00000011198), score: 0.47 ACAD9acyl-CoA dehydrogenase family, member 9 (ENSG00000177646), score: 0.4 ACE2angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 (ENSG00000130234), score: 0.38 ACSL5acyl-CoA synthetase long-chain family member 5 (ENSG00000197142), score: 0.46 ACSS3acyl-CoA synthetase short-chain family member 3 (ENSG00000111058), score: 0.36 ACVR1Cactivin A receptor, type IC (ENSG00000123612), score: 0.37 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.57 ADIPOQadiponectin, C1Q and collagen domain containing (ENSG00000181092), score: 0.38 ADSLadenylosuccinate lyase (ENSG00000239900), score: 0.62 AFMIDarylformamidase (ENSG00000183077), score: 0.41 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.43 AKAP12A kinase (PRKA) anchor protein 12 (ENSG00000131016), score: -0.54 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.47 AKT3v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma) (ENSG00000117020), score: -0.43 ALBalbumin (ENSG00000163631), score: 0.5 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.6 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: 0.37 AMBPalpha-1-microglobulin/bikunin precursor (ENSG00000106927), score: 0.47 AMDHD1amidohydrolase domain containing 1 (ENSG00000139344), score: 0.54 AMOTL1angiomotin like 1 (ENSG00000166025), score: -0.48 ANKRD27ankyrin repeat domain 27 (VPS9 domain) (ENSG00000105186), score: -0.44 AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.49 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: 0.54 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (ENSG00000163697), score: -0.51 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (ENSG00000113108), score: -0.46 ARCN1archain 1 (ENSG00000095139), score: 0.43 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: 0.39 ARL3ADP-ribosylation factor-like 3 (ENSG00000138175), score: -0.49 ARL5BADP-ribosylation factor-like 5B (ENSG00000165997), score: 0.48 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ENSG00000153317), score: -0.44 ASPGasparaginase homolog (S. cerevisiae) (ENSG00000166183), score: 0.42 ATL2atlastin GTPase 2 (ENSG00000119787), score: 0.36 BACH1BTB and CNC homology 1, basic leucine zipper transcription factor 1 (ENSG00000156273), score: 0.37 BAMBIBMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (ENSG00000095739), score: 0.36 BAT2L1HLA-B associated transcript 2-like 1 (ENSG00000130723), score: -0.42 BATF3basic leucine zipper transcription factor, ATF-like 3 (ENSG00000123685), score: 0.39 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (ENSG00000083123), score: 0.38 BICD2bicaudal D homolog 2 (Drosophila) (ENSG00000185963), score: -0.41 BLMHbleomycin hydrolase (ENSG00000108578), score: 0.45 BTKBruton agammaglobulinemia tyrosine kinase (ENSG00000010671), score: 0.43 C13orf15chromosome 13 open reading frame 15 (ENSG00000102760), score: -0.47 C14orf1chromosome 14 open reading frame 1 (ENSG00000133935), score: 0.4 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.45 C14orf179chromosome 14 open reading frame 179 (ENSG00000119650), score: -0.45 C18orf8chromosome 18 open reading frame 8 (ENSG00000141452), score: 0.4 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.36 C5complement component 5 (ENSG00000106804), score: 0.47 C8Acomplement component 8, alpha polypeptide (ENSG00000157131), score: 0.45 C8Bcomplement component 8, beta polypeptide (ENSG00000021852), score: 0.5 C8Gcomplement component 8, gamma polypeptide (ENSG00000176919), score: 0.4 CA6carbonic anhydrase VI (ENSG00000131686), score: 1 CAPN2calpain 2, (m/II) large subunit (ENSG00000162909), score: -0.57 CBY1chibby homolog 1 (Drosophila) (ENSG00000100211), score: -0.45 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: 0.42 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.49 CD28CD28 molecule (ENSG00000178562), score: 0.49 CD34CD34 molecule (ENSG00000174059), score: -0.44 CDC42BPACDC42 binding protein kinase alpha (DMPK-like) (ENSG00000143776), score: -0.47 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 0.39 CDH1cadherin 1, type 1, E-cadherin (epithelial) (ENSG00000039068), score: 0.44 CDO1cysteine dioxygenase, type I (ENSG00000129596), score: 0.43 CEPT1choline/ethanolamine phosphotransferase 1 (ENSG00000134255), score: 0.59 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.49 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.42 CISHcytokine inducible SH2-containing protein (ENSG00000114737), score: 0.36 CKAP4cytoskeleton-associated protein 4 (ENSG00000136026), score: -0.42 CKAP5cytoskeleton associated protein 5 (ENSG00000175216), score: -0.43 CLCN4chloride channel 4 (ENSG00000073464), score: -0.43 CLDN1claudin 1 (ENSG00000163347), score: 0.52 CLN5ceroid-lipofuscinosis, neuronal 5 (ENSG00000102805), score: 0.41 CLPXClpX caseinolytic peptidase X homolog (E. coli) (ENSG00000166855), score: 0.44 COL4A3BPcollagen, type IV, alpha 3 (Goodpasture antigen) binding protein (ENSG00000113163), score: 0.38 COPB1coatomer protein complex, subunit beta 1 (ENSG00000129083), score: 0.39 CPB2carboxypeptidase B2 (plasma) (ENSG00000080618), score: 0.47 CRLS1cardiolipin synthase 1 (ENSG00000088766), score: 0.43 CRYABcrystallin, alpha B (ENSG00000109846), score: -0.42 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.44 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.37 CTNNAL1catenin (cadherin-associated protein), alpha-like 1 (ENSG00000119326), score: -0.42 CUEDC2CUE domain containing 2 (ENSG00000107874), score: -0.59 CXXC5CXXC finger 5 (ENSG00000171604), score: -0.61 CYB5R2cytochrome b5 reductase 2 (ENSG00000166394), score: 0.46 CYB5R3cytochrome b5 reductase 3 (ENSG00000100243), score: -0.43 CYBASC3cytochrome b, ascorbate dependent 3 (ENSG00000162144), score: 0.36 CYBBcytochrome b-245, beta polypeptide (ENSG00000165168), score: 0.44 CYP39A1cytochrome P450, family 39, subfamily A, polypeptide 1 (ENSG00000146233), score: 0.62 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (ENSG00000172817), score: 0.53 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.54 DDOSTdolichyl-diphosphooligosaccharide--protein glycosyltransferase (ENSG00000244038), score: 0.38 DENND1BDENN/MADD domain containing 1B (ENSG00000213047), score: 0.44 DENND5ADENN/MADD domain containing 5A (ENSG00000184014), score: -0.6 DERL1Der1-like domain family, member 1 (ENSG00000136986), score: 0.42 DERL3Der1-like domain family, member 3 (ENSG00000099958), score: 0.4 DHRS11dehydrogenase/reductase (SDR family) member 11 (ENSG00000108272), score: -0.53 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.46 DNAJC22DnaJ (Hsp40) homolog, subfamily C, member 22 (ENSG00000178401), score: 0.39 DNAJC3DnaJ (Hsp40) homolog, subfamily C, member 3 (ENSG00000102580), score: 0.42 DPYSdihydropyrimidinase (ENSG00000147647), score: 0.4 DRAM2DNA-damage regulated autophagy modulator 2 (ENSG00000156171), score: 0.44 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.41 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (ENSG00000134109), score: 0.37 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.53 EEDembryonic ectoderm development (ENSG00000074266), score: -0.45 EFNB2ephrin-B2 (ENSG00000125266), score: -0.42 EGFRepidermal growth factor receptor (ENSG00000146648), score: 0.44 EHHADHenoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (ENSG00000113790), score: 0.37 EPB41L3erythrocyte membrane protein band 4.1-like 3 (ENSG00000082397), score: -0.45 EPS15L1epidermal growth factor receptor pathway substrate 15-like 1 (ENSG00000127527), score: -0.42 EPT1ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) (ENSG00000138018), score: 0.49 EREGepiregulin (ENSG00000124882), score: 0.45 ERGIC2ERGIC and golgi 2 (ENSG00000087502), score: 0.45 ERLEC1endoplasmic reticulum lectin 1 (ENSG00000068912), score: 0.55 ESR1estrogen receptor 1 (ENSG00000091831), score: 0.51 ETNK2ethanolamine kinase 2 (ENSG00000143845), score: 0.37 EVI5ecotropic viral integration site 5 (ENSG00000067208), score: 0.43 F10coagulation factor X (ENSG00000126218), score: 0.55 F2coagulation factor II (thrombin) (ENSG00000180210), score: 0.46 F3coagulation factor III (thromboplastin, tissue factor) (ENSG00000117525), score: -0.42 F9coagulation factor IX (ENSG00000101981), score: 0.55 FAF2Fas associated factor family member 2 (ENSG00000113194), score: 0.53 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.44 FAM167Bfamily with sequence similarity 167, member B (ENSG00000183615), score: 0.4 FAM20Afamily with sequence similarity 20, member A (ENSG00000108950), score: 0.5 FAM3Cfamily with sequence similarity 3, member C (ENSG00000196937), score: -0.53 FAM69Bfamily with sequence similarity 69, member B (ENSG00000165716), score: -0.45 FBXL20F-box and leucine-rich repeat protein 20 (ENSG00000108306), score: 0.49 FETUBfetuin B (ENSG00000090512), score: 0.58 FGBfibrinogen beta chain (ENSG00000171564), score: 0.45 FGF19fibroblast growth factor 19 (ENSG00000162344), score: 0.43 FGFR4fibroblast growth factor receptor 4 (ENSG00000160867), score: 0.36 FGGfibrinogen gamma chain (ENSG00000171557), score: 0.47 FNDC3Afibronectin type III domain containing 3A (ENSG00000102531), score: 0.37 FZD5frizzled homolog 5 (Drosophila) (ENSG00000163251), score: 0.57 GAS6growth arrest-specific 6 (ENSG00000183087), score: -0.55 GAS8growth arrest-specific 8 (ENSG00000141013), score: -0.42 GATA6GATA binding protein 6 (ENSG00000141448), score: 0.38 GCH1GTP cyclohydrolase 1 (ENSG00000131979), score: 0.43 GCHFRGTP cyclohydrolase I feedback regulator (ENSG00000137880), score: 0.38 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.43 GJB1gap junction protein, beta 1, 32kDa (ENSG00000169562), score: 0.36 GLDCglycine dehydrogenase (decarboxylating) (ENSG00000178445), score: 0.38 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: -0.49 GNMTglycine N-methyltransferase (ENSG00000124713), score: 0.45 GOLIM4golgi integral membrane protein 4 (ENSG00000173905), score: 0.5 GPHNgephyrin (ENSG00000171723), score: 0.41 GPLD1glycosylphosphatidylinositol specific phospholipase D1 (ENSG00000112293), score: 0.46 GTPBP2GTP binding protein 2 (ENSG00000172432), score: -0.43 H6PDhexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (ENSG00000049239), score: 0.4 HABP2hyaluronan binding protein 2 (ENSG00000148702), score: 0.41 HALhistidine ammonia-lyase (ENSG00000084110), score: 0.58 HAO1hydroxyacid oxidase (glycolate oxidase) 1 (ENSG00000101323), score: 0.43 HELBhelicase (DNA) B (ENSG00000127311), score: 0.4 HGDhomogentisate 1,2-dioxygenase (ENSG00000113924), score: 0.38 HHIPL1HHIP-like 1 (ENSG00000182218), score: 0.46 HIBCH3-hydroxyisobutyryl-CoA hydrolase (ENSG00000198130), score: 0.41 HM13histocompatibility (minor) 13 (ENSG00000101294), score: 0.37 HNF4Ghepatocyte nuclear factor 4, gamma (ENSG00000164749), score: 0.45 HPGDShematopoietic prostaglandin D synthase (ENSG00000163106), score: 0.51 HPXhemopexin (ENSG00000110169), score: 0.5 HRGhistidine-rich glycoprotein (ENSG00000113905), score: 0.43 HTThuntingtin (ENSG00000197386), score: -0.45 IDH1isocitrate dehydrogenase 1 (NADP+), soluble (ENSG00000138413), score: 0.36 IHHIndian hedgehog (ENSG00000163501), score: 0.46 IKZF3IKAROS family zinc finger 3 (Aiolos) (ENSG00000161405), score: 0.37 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.63 IL15interleukin 15 (ENSG00000164136), score: 0.52 IL22RA2interleukin 22 receptor, alpha 2 (ENSG00000164485), score: 0.83 IL5RAinterleukin 5 receptor, alpha (ENSG00000091181), score: 0.42 IL7Rinterleukin 7 receptor (ENSG00000168685), score: 0.39 IRF8interferon regulatory factor 8 (ENSG00000140968), score: 0.44 ITGA4integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) (ENSG00000115232), score: 0.44 IYDiodotyrosine deiodinase (ENSG00000009765), score: 0.56 KCTD6potassium channel tetramerisation domain containing 6 (ENSG00000168301), score: 0.42 KIAA0090KIAA0090 (ENSG00000127463), score: 0.38 KIAA0427KIAA0427 (ENSG00000134030), score: -0.47 KIAA0430KIAA0430 (ENSG00000166783), score: -0.44 KIAA1432KIAA1432 (ENSG00000107036), score: 0.39 KIAA1958KIAA1958 (ENSG00000165185), score: 0.41 KLHDC8Bkelch domain containing 8B (ENSG00000185909), score: -0.5 KLHL25kelch-like 25 (Drosophila) (ENSG00000183655), score: 0.47 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (ENSG00000117009), score: 0.44 LACTBlactamase, beta (ENSG00000103642), score: 0.46 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.59 LEAP2liver expressed antimicrobial peptide 2 (ENSG00000164406), score: 0.5 LEPROTL1leptin receptor overlapping transcript-like 1 (ENSG00000104660), score: -0.43 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.64 LIPClipase, hepatic (ENSG00000166035), score: 0.36 LOC100292021similar to thioredoxin peroxidase (ENSG00000123131), score: 0.53 LPGAT1lysophosphatidylglycerol acyltransferase 1 (ENSG00000123684), score: 0.38 LRP11low density lipoprotein receptor-related protein 11 (ENSG00000120256), score: -0.5 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.74 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.66 LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.48 LY75lymphocyte antigen 75 (ENSG00000054219), score: 0.38 LYNv-yes-1 Yamaguchi sarcoma viral related oncogene homolog (ENSG00000147507), score: 0.43 LZTS2leucine zipper, putative tumor suppressor 2 (ENSG00000107816), score: -0.41 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.42 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (ENSG00000138834), score: -0.46 MARVELD2MARVEL domain containing 2 (ENSG00000152939), score: 0.44 MASP1mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) (ENSG00000127241), score: 0.37 MASP2mannan-binding lectin serine peptidase 2 (ENSG00000009724), score: 0.39 MAST4microtubule associated serine/threonine kinase family member 4 (ENSG00000069020), score: -0.45 MAT1Amethionine adenosyltransferase I, alpha (ENSG00000151224), score: 0.49 MATN2matrilin 2 (ENSG00000132561), score: -0.45 MBD3methyl-CpG binding domain protein 3 (ENSG00000071655), score: -0.49 MBNL2muscleblind-like 2 (Drosophila) (ENSG00000139793), score: -0.44 MFSD2Amajor facilitator superfamily domain containing 2A (ENSG00000168389), score: 0.38 MKLN1muskelin 1, intracellular mediator containing kelch motifs (ENSG00000128585), score: 0.62 MLLT1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 (ENSG00000130382), score: -0.44 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.52 MPDZmultiple PDZ domain protein (ENSG00000107186), score: -0.48 MRPL44mitochondrial ribosomal protein L44 (ENSG00000135900), score: 0.39 MTA1metastasis associated 1 (ENSG00000182979), score: -0.63 MTMR2myotubularin related protein 2 (ENSG00000087053), score: -0.44 MTPAPmitochondrial poly(A) polymerase (ENSG00000107951), score: 0.44 MTTPmicrosomal triglyceride transfer protein (ENSG00000138823), score: 0.49 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: 0.51 NAPRT1nicotinate phosphoribosyltransferase domain containing 1 (ENSG00000147813), score: 0.36 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: 0.4 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.39 NR5A2nuclear receptor subfamily 5, group A, member 2 (ENSG00000116833), score: 0.48 NRBP1nuclear receptor binding protein 1 (ENSG00000115216), score: -0.55 NTSR1neurotensin receptor 1 (high affinity) (ENSG00000101188), score: 0.44 NUP93nucleoporin 93kDa (ENSG00000102900), score: -0.44 OIT3oncoprotein induced transcript 3 (ENSG00000138315), score: 0.48 ONECUT1one cut homeobox 1 (ENSG00000169856), score: 0.48 OSTalphaorganic solute transporter alpha (ENSG00000163959), score: 0.41 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (ENSG00000122884), score: 0.39 PACRGLPARK2 co-regulated-like (ENSG00000163138), score: -0.45 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.46 PAHphenylalanine hydroxylase (ENSG00000171759), score: 0.4 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (ENSG00000149269), score: -0.42 PALMparalemmin (ENSG00000099864), score: -0.45 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.47 PAPSS23'-phosphoadenosine 5'-phosphosulfate synthase 2 (ENSG00000198682), score: 0.36 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.37 PARP16poly (ADP-ribose) polymerase family, member 16 (ENSG00000138617), score: 0.36 PARP6poly (ADP-ribose) polymerase family, member 6 (ENSG00000137817), score: -0.5 PATZ1POZ (BTB) and AT hook containing zinc finger 1 (ENSG00000100105), score: -0.41 PCGF2polycomb group ring finger 2 (ENSG00000056661), score: -0.43 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.46 PDCD1LG2programmed cell death 1 ligand 2 (ENSG00000197646), score: 0.42 PDE1Aphosphodiesterase 1A, calmodulin-dependent (ENSG00000115252), score: -0.41 PDE6Dphosphodiesterase 6D, cGMP-specific, rod, delta (ENSG00000156973), score: -0.5 PDIA3protein disulfide isomerase family A, member 3 (ENSG00000167004), score: 0.42 PDIK1LPDLIM1 interacting kinase 1 like (ENSG00000175087), score: 0.37 PEMTphosphatidylethanolamine N-methyltransferase (ENSG00000133027), score: 0.4 PEX13peroxisomal biogenesis factor 13 (ENSG00000162928), score: 0.39 PHF16PHD finger protein 16 (ENSG00000102221), score: 0.46 PIGXphosphatidylinositol glycan anchor biosynthesis, class X (ENSG00000163964), score: 0.39 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (ENSG00000186111), score: -0.42 PLA2G12Bphospholipase A2, group XIIB (ENSG00000138308), score: 0.45 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.37 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (ENSG00000125630), score: 0.38 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (ENSG00000132664), score: 0.39 PPIBpeptidylprolyl isomerase B (cyclophilin B) (ENSG00000166794), score: 0.36 PPP1R8protein phosphatase 1, regulatory (inhibitor) subunit 8 (ENSG00000117751), score: -0.46 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta (ENSG00000175470), score: -0.45 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta (ENSG00000112640), score: -0.56 PROZprotein Z, vitamin K-dependent plasma glycoprotein (ENSG00000126231), score: 0.44 PSMA3proteasome (prosome, macropain) subunit, alpha type, 3 (ENSG00000100567), score: 0.4 PTPRGprotein tyrosine phosphatase, receptor type, G (ENSG00000144724), score: 0.51 PYGLphosphorylase, glycogen, liver (ENSG00000100504), score: 0.4 QPCTLglutaminyl-peptide cyclotransferase-like (ENSG00000011478), score: 0.41 RAB30RAB30, member RAS oncogene family (ENSG00000137502), score: 0.4 RAB33BRAB33B, member RAS oncogene family (ENSG00000172007), score: 0.42 RANBP10RAN binding protein 10 (ENSG00000141084), score: 0.4 RBP4retinol binding protein 4, plasma (ENSG00000138207), score: 0.4 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.43 RP2retinitis pigmentosa 2 (X-linked recessive) (ENSG00000102218), score: 0.37 RPIAribose 5-phosphate isomerase A (ENSG00000153574), score: 0.43 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (ENSG00000211456), score: 0.36 SAR1BSAR1 homolog B (S. cerevisiae) (ENSG00000152700), score: 0.52 SASH3SAM and SH3 domain containing 3 (ENSG00000122122), score: 0.47 SCUBE3signal peptide, CUB domain, EGF-like 3 (ENSG00000146197), score: 0.4 SDC2syndecan 2 (ENSG00000169439), score: 0.4 SDCCAG8serologically defined colon cancer antigen 8 (ENSG00000054282), score: -0.45 SDR42E1short chain dehydrogenase/reductase family 42E, member 1 (ENSG00000184860), score: 0.53 SEBOXSEBOX homeobox (ENSG00000109072), score: 0.46 SEC23ASec23 homolog A (S. cerevisiae) (ENSG00000100934), score: 0.41 SERPINA10serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 (ENSG00000140093), score: 0.48 SERPINC1serpin peptidase inhibitor, clade C (antithrombin), member 1 (ENSG00000117601), score: 0.47 SESN1sestrin 1 (ENSG00000080546), score: 0.46 SETD5SET domain containing 5 (ENSG00000168137), score: -0.45 SGCEsarcoglycan, epsilon (ENSG00000127990), score: -0.66 SGPL1sphingosine-1-phosphate lyase 1 (ENSG00000166224), score: 0.37 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: -0.56 SIKE1suppressor of IKBKE 1 (ENSG00000052723), score: 0.41 SIPA1L1signal-induced proliferation-associated 1 like 1 (ENSG00000197555), score: -0.42 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (ENSG00000155380), score: 0.36 SLC25A38solute carrier family 25, member 38 (ENSG00000144659), score: -0.41 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.57 SLC2A2solute carrier family 2 (facilitated glucose transporter), member 2 (ENSG00000163581), score: 0.4 SLC2A9solute carrier family 2 (facilitated glucose transporter), member 9 (ENSG00000109667), score: 0.5 SLC30A1solute carrier family 30 (zinc transporter), member 1 (ENSG00000170385), score: 0.57 SLC30A10solute carrier family 30, member 10 (ENSG00000196660), score: 0.5 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.37 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.54 SLC31A1solute carrier family 31 (copper transporters), member 1 (ENSG00000136868), score: 0.48 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (ENSG00000169359), score: 0.4 SLC37A1solute carrier family 37 (glycerol-3-phosphate transporter), member 1 (ENSG00000160190), score: -0.41 SLC38A4solute carrier family 38, member 4 (ENSG00000139209), score: 0.52 SLC39A8solute carrier family 39 (zinc transporter), member 8 (ENSG00000138821), score: 0.37 SLC39A9solute carrier family 39 (zinc transporter), member 9 (ENSG00000029364), score: 0.53 SLC7A2solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 (ENSG00000003989), score: 0.36 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (ENSG00000103064), score: -0.42 SLIT3slit homolog 3 (Drosophila) (ENSG00000184347), score: -0.48 SNX24sorting nexin 24 (ENSG00000064652), score: 0.4 SNX33sorting nexin 33 (ENSG00000173548), score: 0.38 SPI1spleen focus forming virus (SFFV) proviral integration oncogene spi1 (ENSG00000066336), score: 0.4 SPINK4serine peptidase inhibitor, Kazal type 4 (ENSG00000122711), score: 0.69 SPIRE1spire homolog 1 (Drosophila) (ENSG00000134278), score: -0.47 SPON1spondin 1, extracellular matrix protein (ENSG00000152268), score: -0.46 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.6 SPPL2Bsignal peptide peptidase-like 2B (ENSG00000005206), score: -0.45 SRSF4serine/arginine-rich splicing factor 4 (ENSG00000116350), score: -0.6 SSR1signal sequence receptor, alpha (ENSG00000124783), score: 0.55 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: 0.41 ST14suppression of tumorigenicity 14 (colon carcinoma) (ENSG00000149418), score: 0.38 STARD5StAR-related lipid transfer (START) domain containing 5 (ENSG00000172345), score: 0.38 STAT3signal transducer and activator of transcription 3 (acute-phase response factor) (ENSG00000168610), score: 0.38 STK24serine/threonine kinase 24 (ENSG00000102572), score: -0.51 STK39serine threonine kinase 39 (ENSG00000198648), score: -0.45 STOML3stomatin (EPB72)-like 3 (ENSG00000133115), score: 0.67 SYDE2synapse defective 1, Rho GTPase, homolog 2 (C. elegans) (ENSG00000097096), score: 0.43 TAAR1trace amine associated receptor 1 (ENSG00000146399), score: 0.65 TAB3TGF-beta activated kinase 1/MAP3K7 binding protein 3 (ENSG00000157625), score: 0.36 TADA2Btranscriptional adaptor 2B (ENSG00000173011), score: -0.49 TAPT1transmembrane anterior posterior transformation 1 (ENSG00000169762), score: 0.43 TATtyrosine aminotransferase (ENSG00000198650), score: 0.48 TATDN1TatD DNase domain containing 1 (ENSG00000147687), score: 0.41 TBX3T-box 3 (ENSG00000135111), score: 0.38 TDO2tryptophan 2,3-dioxygenase (ENSG00000151790), score: 0.45 THUMPD2THUMP domain containing 2 (ENSG00000138050), score: 0.46 TLR3toll-like receptor 3 (ENSG00000164342), score: 0.68 TM4SF4transmembrane 4 L six family member 4 (ENSG00000169903), score: 0.54 TM7SF4transmembrane 7 superfamily member 4 (ENSG00000164935), score: 0.54 TMCO3transmembrane and coiled-coil domains 3 (ENSG00000150403), score: -0.45 TMED5transmembrane emp24 protein transport domain containing 5 (ENSG00000117500), score: 0.39 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.5 TMEM195transmembrane protein 195 (ENSG00000187546), score: 0.62 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.38 TMEM41Btransmembrane protein 41B (ENSG00000166471), score: 0.51 TMEM82transmembrane protein 82 (ENSG00000162460), score: 0.36 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.48 TOM1L1target of myb1 (chicken)-like 1 (ENSG00000141198), score: -0.41 TP53INP1tumor protein p53 inducible nuclear protein 1 (ENSG00000164938), score: 0.39 TPST1tyrosylprotein sulfotransferase 1 (ENSG00000169902), score: 0.48 TRAF3IP3TRAF3 interacting protein 3 (ENSG00000009790), score: 0.43 TRPA1transient receptor potential cation channel, subfamily A, member 1 (ENSG00000104321), score: 0.5 TSHBthyroid stimulating hormone, beta (ENSG00000134200), score: 0.37 TSPAN15tetraspanin 15 (ENSG00000099282), score: -0.51 TTC1tetratricopeptide repeat domain 1 (ENSG00000113312), score: -0.46 TTC36tetratricopeptide repeat domain 36 (ENSG00000172425), score: 0.39 TTC39Btetratricopeptide repeat domain 39B (ENSG00000155158), score: 0.37 TTPAtocopherol (alpha) transfer protein (ENSG00000137561), score: 0.61 TWISTNBTWIST neighbor (ENSG00000105849), score: 0.37 TWSG1twisted gastrulation homolog 1 (Drosophila) (ENSG00000128791), score: -0.59 UBXN4UBX domain protein 4 (ENSG00000144224), score: 0.43 UNC119unc-119 homolog (C. elegans) (ENSG00000109103), score: -0.57 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: 0.6 USP54ubiquitin specific peptidase 54 (ENSG00000166348), score: -0.42 UTP15UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) (ENSG00000164338), score: 0.39 VSIG4V-set and immunoglobulin domain containing 4 (ENSG00000155659), score: 0.5 WDFY2WD repeat and FYVE domain containing 2 (ENSG00000139668), score: 0.53 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.43 WDR60WD repeat domain 60 (ENSG00000126870), score: -0.41 WDR8WD repeat domain 8 (ENSG00000116213), score: -0.45 WLSwntless homolog (Drosophila) (ENSG00000116729), score: -0.5 XKR9XK, Kell blood group complex subunit-related family, member 9 (ENSG00000221947), score: 0.42 XRCC6BP1XRCC6 binding protein 1 (ENSG00000166896), score: 0.45 YIPF5Yip1 domain family, member 5 (ENSG00000145817), score: 0.58 ZBTB2zinc finger and BTB domain containing 2 (ENSG00000181472), score: 0.43 ZC3H12Dzinc finger CCCH-type containing 12D (ENSG00000178199), score: 0.63 ZCCHC11zinc finger, CCHC domain containing 11 (ENSG00000134744), score: 0.41 ZMPSTE24zinc metallopeptidase (STE24 homolog, S. cerevisiae) (ENSG00000084073), score: 0.4 ZMYND8zinc finger, MYND-type containing 8 (ENSG00000101040), score: -0.43 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.39 ZNF462zinc finger protein 462 (ENSG00000148143), score: -0.47 ZP1zona pellucida glycoprotein 1 (sperm receptor) (ENSG00000149506), score: 0.42
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_lv_f_ca1 | gga | lv | f | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |
| gga_lv_m_ca1 | gga | lv | m | _ |