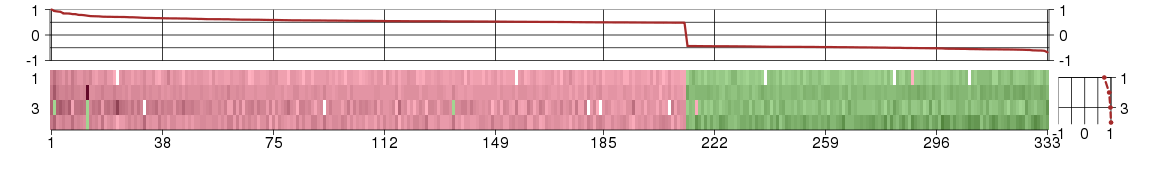

Under-expression is coded with green,
over-expression with red color.



ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: -0.55 ABL1c-abl oncogene 1, non-receptor tyrosine kinase (ENSG00000097007), score: 0.5 ACP6acid phosphatase 6, lysophosphatidic (ENSG00000162836), score: -0.51 ADAMTS14ADAM metallopeptidase with thrombospondin type 1 motif, 14 (ENSG00000138316), score: 0.71 ADAMTS8ADAM metallopeptidase with thrombospondin type 1 motif, 8 (ENSG00000134917), score: 0.54 ADARB2adenosine deaminase, RNA-specific, B2 (ENSG00000185736), score: 0.66 AKAP9A kinase (PRKA) anchor protein (yotiao) 9 (ENSG00000127914), score: -0.51 ALKBH8alkB, alkylation repair homolog 8 (E. coli) (ENSG00000137760), score: -0.49 ANO4anoctamin 4 (ENSG00000151572), score: 0.55 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: -0.49 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: -0.47 ARHGAP1Rho GTPase activating protein 1 (ENSG00000175220), score: 0.53 ARHGAP22Rho GTPase activating protein 22 (ENSG00000128805), score: 0.58 ARL8AADP-ribosylation factor-like 8A (ENSG00000143862), score: 0.53 ARRDC2arrestin domain containing 2 (ENSG00000105643), score: 0.53 ATL2atlastin GTPase 2 (ENSG00000119787), score: -0.68 ATP11AATPase, class VI, type 11A (ENSG00000068650), score: 0.49 ATP13A2ATPase type 13A2 (ENSG00000159363), score: 0.51 BACE1beta-site APP-cleaving enzyme 1 (ENSG00000186318), score: 0.54 BCAS1breast carcinoma amplified sequence 1 (ENSG00000064787), score: 0.55 BCAS3breast carcinoma amplified sequence 3 (ENSG00000141376), score: 0.62 BTBD2BTB (POZ) domain containing 2 (ENSG00000133243), score: 0.5 C12orf45chromosome 12 open reading frame 45 (ENSG00000151131), score: -0.46 C16orf42chromosome 16 open reading frame 42 (ENSG00000007520), score: 0.56 C16orf70chromosome 16 open reading frame 70 (ENSG00000125149), score: 0.54 C1orf128chromosome 1 open reading frame 128 (ENSG00000057757), score: 0.5 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.71 C5orf41chromosome 5 open reading frame 41 (ENSG00000164463), score: -0.46 C8orf38chromosome 8 open reading frame 38 (ENSG00000156170), score: -0.47 C8orf46chromosome 8 open reading frame 46 (ENSG00000169085), score: 0.54 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 0.94 CAPN13calpain 13 (ENSG00000162949), score: 0.49 CAPN9calpain 9 (ENSG00000135773), score: 0.84 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: -0.52 CCM2cerebral cavernous malformation 2 (ENSG00000136280), score: 0.52 CD2APCD2-associated protein (ENSG00000198087), score: -0.48 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: 0.59 CDH20cadherin 20, type 2 (ENSG00000101542), score: 0.53 CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.57 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (ENSG00000115816), score: -0.45 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 1 CETN3centrin, EF-hand protein, 3 (ENSG00000153140), score: -0.47 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: -0.43 CIITAclass II, major histocompatibility complex, transactivator (ENSG00000179583), score: 0.51 CLCN6chloride channel 6 (ENSG00000011021), score: 0.49 CLCN7chloride channel 7 (ENSG00000103249), score: 0.55 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.92 CNDP1carnosine dipeptidase 1 (metallopeptidase M20 family) (ENSG00000150656), score: 0.54 CNTN2contactin 2 (axonal) (ENSG00000184144), score: 0.63 COBRA1cofactor of BRCA1 (ENSG00000188986), score: 0.49 COG1component of oligomeric golgi complex 1 (ENSG00000166685), score: 0.57 COL4A1collagen, type IV, alpha 1 (ENSG00000187498), score: -0.44 COMMD10COMM domain containing 10 (ENSG00000145781), score: -0.48 COMMD2COMM domain containing 2 (ENSG00000114744), score: -0.45 COX10COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) (ENSG00000006695), score: -0.5 CPNE2copine II (ENSG00000140848), score: 0.62 CSRP1cysteine and glycine-rich protein 1 (ENSG00000159176), score: 0.57 CTBP1C-terminal binding protein 1 (ENSG00000159692), score: 0.5 CTNNAL1catenin (cadherin-associated protein), alpha-like 1 (ENSG00000119326), score: -0.48 CTRLchymotrypsin-like (ENSG00000141086), score: 0.55 CX3CR1chemokine (C-X3-C motif) receptor 1 (ENSG00000168329), score: 0.49 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: -0.48 DAAM2dishevelled associated activator of morphogenesis 2 (ENSG00000146122), score: 0.7 DDX46DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 (ENSG00000145833), score: -0.47 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (ENSG00000141141), score: -0.56 DFNB31deafness, autosomal recessive 31 (ENSG00000095397), score: 0.54 DGCR2DiGeorge syndrome critical region gene 2 (ENSG00000070413), score: 0.52 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: -0.44 DNMBPdynamin binding protein (ENSG00000107554), score: -0.62 DOHHdeoxyhypusine hydroxylase/monooxygenase (ENSG00000129932), score: 0.65 DPYSL4dihydropyrimidinase-like 4 (ENSG00000151640), score: 0.51 DSCAML1Down syndrome cell adhesion molecule like 1 (ENSG00000177103), score: 0.58 DSELdermatan sulfate epimerase-like (ENSG00000171451), score: 0.58 DYNC1I2dynein, cytoplasmic 1, intermediate chain 2 (ENSG00000077380), score: 0.72 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (ENSG00000134001), score: -0.47 EIF3Eeukaryotic translation initiation factor 3, subunit E (ENSG00000104408), score: -0.49 EIF3Meukaryotic translation initiation factor 3, subunit M (ENSG00000149100), score: -0.52 EIF4Heukaryotic translation initiation factor 4H (ENSG00000106682), score: 0.59 EIF5Beukaryotic translation initiation factor 5B (ENSG00000158417), score: -0.48 ELOVL1elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (ENSG00000066322), score: 0.7 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENSG00000136960), score: 0.57 ENTPD6ectonucleoside triphosphate diphosphohydrolase 6 (putative) (ENSG00000197586), score: 0.54 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: -0.54 EPN2epsin 2 (ENSG00000072134), score: 0.53 ERI1exoribonuclease 1 (ENSG00000104626), score: -0.45 ERMNermin, ERM-like protein (ENSG00000136541), score: 0.59 EVI2Aecotropic viral integration site 2A (ENSG00000126860), score: 0.71 EXOC7exocyst complex component 7 (ENSG00000182473), score: 0.6 FA2Hfatty acid 2-hydroxylase (ENSG00000103089), score: 0.64 FAM102Afamily with sequence similarity 102, member A (ENSG00000167106), score: 0.73 FAM124Afamily with sequence similarity 124A (ENSG00000150510), score: 0.79 FAM171A1family with sequence similarity 171, member A1 (ENSG00000148468), score: 0.54 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: -0.55 FAM69Cfamily with sequence similarity 69, member C (ENSG00000187773), score: 0.63 FASTKD2FAST kinase domains 2 (ENSG00000118246), score: -0.48 FBXO2F-box protein 2 (ENSG00000116661), score: 0.5 FBXW4F-box and WD repeat domain containing 4 (ENSG00000107829), score: 0.48 FCHO2FCH domain only 2 (ENSG00000157107), score: -0.44 FEZF2FEZ family zinc finger 2 (ENSG00000153266), score: 0.54 FGF22fibroblast growth factor 22 (ENSG00000070388), score: 0.65 FGFR2fibroblast growth factor receptor 2 (ENSG00000066468), score: 0.5 FKBP15FK506 binding protein 15, 133kDa (ENSG00000119321), score: 0.54 FNBP1Lformin binding protein 1-like (ENSG00000137942), score: -0.58 FOXG1forkhead box G1 (ENSG00000176165), score: 0.52 FZD6frizzled homolog 6 (Drosophila) (ENSG00000164930), score: -0.53 GJD4gap junction protein, delta 4, 40.1kDa (ENSG00000177291), score: 0.67 GLDNgliomedin (ENSG00000186417), score: 0.82 GLT25D2glycosyltransferase 25 domain containing 2 (ENSG00000198756), score: 0.52 GNAZguanine nucleotide binding protein (G protein), alpha z polypeptide (ENSG00000128266), score: 0.49 GNL2guanine nucleotide binding protein-like 2 (nucleolar) (ENSG00000134697), score: -0.54 GNPTGN-acetylglucosamine-1-phosphate transferase, gamma subunit (ENSG00000090581), score: 0.54 GOLGA4golgin A4 (ENSG00000144674), score: -0.57 GORABgolgin, RAB6-interacting (ENSG00000120370), score: -0.54 GPM6Bglycoprotein M6B (ENSG00000046653), score: 0.5 GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.72 GRID1glutamate receptor, ionotropic, delta 1 (ENSG00000182771), score: 0.6 GRIK4glutamate receptor, ionotropic, kainate 4 (ENSG00000149403), score: 0.49 GRK6G protein-coupled receptor kinase 6 (ENSG00000198055), score: 0.49 GRM3glutamate receptor, metabotropic 3 (ENSG00000198822), score: 0.57 GTF2A2general transcription factor IIA, 2, 12kDa (ENSG00000140307), score: -0.44 GTF2E2general transcription factor IIE, polypeptide 2, beta 34kDa (ENSG00000197265), score: -0.55 HDAC11histone deacetylase 11 (ENSG00000163517), score: 0.57 HERC4hect domain and RLD 4 (ENSG00000148634), score: -0.44 HGShepatocyte growth factor-regulated tyrosine kinase substrate (ENSG00000185359), score: 0.57 HHATLhedgehog acyltransferase-like (ENSG00000010282), score: 0.49 HHIPhedgehog interacting protein (ENSG00000164161), score: 0.57 HHIPL1HHIP-like 1 (ENSG00000182218), score: 0.61 HOOK1hook homolog 1 (Drosophila) (ENSG00000134709), score: -0.46 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.67 IDEinsulin-degrading enzyme (ENSG00000119912), score: -0.48 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.62 ING3inhibitor of growth family, member 3 (ENSG00000071243), score: -0.49 INTS8integrator complex subunit 8 (ENSG00000164941), score: -0.56 ITGB4integrin, beta 4 (ENSG00000132470), score: 0.48 ITPK1inositol 1,3,4-triphosphate 5/6 kinase (ENSG00000100605), score: 0.63 JAK2Janus kinase 2 (ENSG00000096968), score: -0.44 JAM3junctional adhesion molecule 3 (ENSG00000166086), score: 0.67 KCNF1potassium voltage-gated channel, subfamily F, member 1 (ENSG00000162975), score: 0.6 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 0.91 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (ENSG00000135643), score: 0.57 KIAA0020KIAA0020 (ENSG00000080608), score: -0.47 KIAA1429KIAA1429 (ENSG00000164944), score: -0.49 KIAA1598KIAA1598 (ENSG00000187164), score: 0.53 KIF13Bkinesin family member 13B (ENSG00000197892), score: 0.52 KIRREL3kin of IRRE like 3 (Drosophila) (ENSG00000149571), score: 0.49 KLHL26kelch-like 26 (Drosophila) (ENSG00000167487), score: 0.56 L3MBTL2l(3)mbt-like 2 (Drosophila) (ENSG00000100395), score: 0.53 LAPTM4Alysosomal protein transmembrane 4 alpha (ENSG00000068697), score: -0.59 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.6 LIMK1LIM domain kinase 1 (ENSG00000106683), score: 0.49 LIN7Clin-7 homolog C (C. elegans) (ENSG00000148943), score: -0.51 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.74 LOC100131693eukaryotic translation initiation factor 4E pseudogene (ENSG00000151247), score: -0.57 LOC100294412similar to KIAA0655 protein (ENSG00000130787), score: 0.66 LOC652522similar to Werner syndrome protein (ENSG00000165392), score: -0.49 LPCAT3lysophosphatidylcholine acyltransferase 3 (ENSG00000111684), score: -0.43 LPPR3lipid phosphate phosphatase-related protein type 3 (ENSG00000129951), score: 0.54 LRCH1leucine-rich repeats and calponin homology (CH) domain containing 1 (ENSG00000136141), score: -0.5 LRFN2leucine rich repeat and fibronectin type III domain containing 2 (ENSG00000156564), score: 0.5 LRIT2leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 (ENSG00000204033), score: 0.53 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (ENSG00000160285), score: 0.52 LTV1LTV1 homolog (S. cerevisiae) (ENSG00000135521), score: -0.51 LYSMD3LysM, putative peptidoglycan-binding, domain containing 3 (ENSG00000176018), score: -0.45 LZTS2leucine zipper, putative tumor suppressor 2 (ENSG00000107816), score: 0.57 MALmal, T-cell differentiation protein (ENSG00000172005), score: 0.54 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: -0.54 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (ENSG00000011566), score: -0.52 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (ENSG00000138834), score: 0.5 MAPKSP1MAPK scaffold protein 1 (ENSG00000109270), score: -0.45 MATN1matrilin 1, cartilage matrix protein (ENSG00000162510), score: 0.58 METTL14methyltransferase like 14 (ENSG00000145388), score: -0.43 MFSD2Bmajor facilitator superfamily domain containing 2B (ENSG00000205639), score: 0.5 MFSD6major facilitator superfamily domain containing 6 (ENSG00000151690), score: 0.49 MGRN1mahogunin, ring finger 1 (ENSG00000102858), score: 0.49 MMAAmethylmalonic aciduria (cobalamin deficiency) cblA type (ENSG00000151611), score: -0.47 MNAT1menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) (ENSG00000020426), score: -0.6 MOBKL2BMOB1, Mps One Binder kinase activator-like 2B (yeast) (ENSG00000120162), score: 0.48 MOCS1molybdenum cofactor synthesis 1 (ENSG00000124615), score: 0.56 MPLmyeloproliferative leukemia virus oncogene (ENSG00000117400), score: 0.51 MRPL15mitochondrial ribosomal protein L15 (ENSG00000137547), score: -0.48 MRPL50mitochondrial ribosomal protein L50 (ENSG00000136897), score: -0.52 MSGN1mesogenin 1 (ENSG00000151379), score: 0.76 MTERFD1MTERF domain containing 1 (ENSG00000156469), score: -0.54 MTMR10myotubularin related protein 10 (ENSG00000166912), score: 0.56 MTPAPmitochondrial poly(A) polymerase (ENSG00000107951), score: -0.46 MVDmevalonate (diphospho) decarboxylase (ENSG00000167508), score: 0.53 MYBPC1myosin binding protein C, slow type (ENSG00000196091), score: 0.49 MYNNmyoneurin (ENSG00000085274), score: -0.53 MYO10myosin X (ENSG00000145555), score: 0.56 MYO1Dmyosin ID (ENSG00000176658), score: 0.48 MYO9Bmyosin IXB (ENSG00000099331), score: 0.66 NDPNorrie disease (pseudoglioma) (ENSG00000124479), score: 0.51 NDST1N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 (ENSG00000070614), score: 0.51 NEO1neogenin 1 (ENSG00000067141), score: 0.51 NFASCneurofascin (ENSG00000163531), score: 0.5 NFU1NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) (ENSG00000169599), score: -0.44 NINJ2ninjurin 2 (ENSG00000171840), score: 0.53 NIPA1non imprinted in Prader-Willi/Angelman syndrome 1 (ENSG00000170113), score: 0.62 NIPAL3NIPA-like domain containing 3 (ENSG00000001461), score: 0.53 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 0.71 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: -0.49 NOTCH1notch 1 (ENSG00000148400), score: 0.5 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.54 NR2C1nuclear receptor subfamily 2, group C, member 1 (ENSG00000120798), score: -0.43 NT5C35'-nucleotidase, cytosolic III (ENSG00000122643), score: -0.47 NUCB2nucleobindin 2 (ENSG00000070081), score: -0.47 OPN4opsin 4 (ENSG00000122375), score: 0.62 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: 0.59 PARP1poly (ADP-ribose) polymerase 1 (ENSG00000143799), score: 0.51 PCDH9protocadherin 9 (ENSG00000184226), score: 0.51 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.7 PEX13peroxisomal biogenesis factor 13 (ENSG00000162928), score: -0.56 PFKLphosphofructokinase, liver (ENSG00000141959), score: 0.49 PHLDA3pleckstrin homology-like domain, family A, member 3 (ENSG00000174307), score: 0.6 PHLDB1pleckstrin homology-like domain, family B, member 1 (ENSG00000019144), score: 0.57 PHLPP1PH domain and leucine rich repeat protein phosphatase 1 (ENSG00000081913), score: 0.6 PIGBphosphatidylinositol glycan anchor biosynthesis, class B (ENSG00000069943), score: -0.52 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (ENSG00000171608), score: 0.55 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.78 PLP1proteolipid protein 1 (ENSG00000123560), score: 0.55 PLXNB1plexin B1 (ENSG00000164050), score: 0.54 PMP2peripheral myelin protein 2 (ENSG00000147588), score: 0.65 POLD3polymerase (DNA-directed), delta 3, accessory subunit (ENSG00000077514), score: -0.49 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (ENSG00000125630), score: -0.46 POMGNT1protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase (ENSG00000085998), score: 0.49 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.79 PRMT10protein arginine methyltransferase 10 (putative) (ENSG00000164169), score: -0.44 PRPH2peripherin 2 (retinal degeneration, slow) (ENSG00000112619), score: 0.54 PRR5Lproline rich 5 like (ENSG00000135362), score: 0.74 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: 0.6 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: 0.73 RAB40CRAB40C, member RAS oncogene family (ENSG00000197562), score: 0.57 RANGAP1Ran GTPase activating protein 1 (ENSG00000100401), score: 0.62 RAPGEF5Rap guanine nucleotide exchange factor (GEF) 5 (ENSG00000136237), score: 0.66 RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.69 RBP3retinol binding protein 3, interstitial (ENSG00000107618), score: 0.71 RFTN2raftlin family member 2 (ENSG00000162944), score: 0.5 RGS11regulator of G-protein signaling 11 (ENSG00000076344), score: 0.53 RHOrhodopsin (ENSG00000163914), score: 0.53 RHOUras homolog gene family, member U (ENSG00000116574), score: 0.54 RHPN1rhophilin, Rho GTPase binding protein 1 (ENSG00000158106), score: 0.5 RIOK2RIO kinase 2 (yeast) (ENSG00000058729), score: -0.6 RNF141ring finger protein 141 (ENSG00000110315), score: 0.49 ROBO3roundabout, axon guidance receptor, homolog 3 (Drosophila) (ENSG00000154134), score: 0.62 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (ENSG00000067900), score: -0.51 RPF1ribosome production factor 1 homolog (S. cerevisiae) (ENSG00000117133), score: -0.61 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (ENSG00000067533), score: -0.54 RS1retinoschisin 1 (ENSG00000102104), score: 0.82 RSPO4R-spondin family, member 4 (ENSG00000101282), score: 0.54 SCRG1stimulator of chondrogenesis 1 (ENSG00000164106), score: 0.53 SECISBP2LSECIS binding protein 2-like (ENSG00000138593), score: 0.59 SEMA6Asema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A (ENSG00000092421), score: 0.69 SETD6SET domain containing 6 (ENSG00000103037), score: -0.57 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.57 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: 0.56 SH3PXD2BSH3 and PX domains 2B (ENSG00000174705), score: 0.5 SIK3SIK family kinase 3 (ENSG00000160584), score: 0.55 SIN3BSIN3 homolog B, transcription regulator (yeast) (ENSG00000127511), score: 0.48 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.51 SLC31A1solute carrier family 31 (copper transporters), member 1 (ENSG00000136868), score: -0.44 SLC31A2solute carrier family 31 (copper transporters), member 2 (ENSG00000136867), score: 0.51 SLC44A1solute carrier family 44, member 1 (ENSG00000070214), score: 0.71 SLC5A11solute carrier family 5 (sodium/glucose cotransporter), member 11 (ENSG00000158865), score: 0.65 SMAD5SMAD family member 5 (ENSG00000113658), score: -0.48 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (ENSG00000100796), score: -0.51 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: -0.57 SNX13sorting nexin 13 (ENSG00000071189), score: -0.46 SNX22sorting nexin 22 (ENSG00000157734), score: 0.51 SOX8SRY (sex determining region Y)-box 8 (ENSG00000005513), score: 0.68 SREBF2sterol regulatory element binding transcription factor 2 (ENSG00000198911), score: 0.52 SSR1signal sequence receptor, alpha (ENSG00000124783), score: -0.47 SSTsomatostatin (ENSG00000157005), score: 0.53 ST7suppression of tumorigenicity 7 (ENSG00000004866), score: -0.46 STARD3StAR-related lipid transfer (START) domain containing 3 (ENSG00000131748), score: 0.57 STK17Bserine/threonine kinase 17b (ENSG00000081320), score: -0.44 STOML1stomatin (EPB72)-like 1 (ENSG00000067221), score: 0.52 STRN3striatin, calmodulin binding protein 3 (ENSG00000196792), score: -0.45 SUZ12suppressor of zeste 12 homolog (Drosophila) (ENSG00000178691), score: -0.61 SYNJ2synaptojanin 2 (ENSG00000078269), score: 0.65 TAF12TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa (ENSG00000120656), score: -0.55 TAF1BTATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa (ENSG00000115750), score: -0.44 TBC1D2TBC1 domain family, member 2 (ENSG00000095383), score: 0.6 TBC1D23TBC1 domain family, member 23 (ENSG00000036054), score: -0.5 TBC1D5TBC1 domain family, member 5 (ENSG00000131374), score: 0.54 TBCEtubulin folding cofactor E (ENSG00000116957), score: -0.47 TDRD3tudor domain containing 3 (ENSG00000083544), score: -0.58 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (ENSG00000114126), score: -0.44 THOC2THO complex 2 (ENSG00000125676), score: -0.58 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: 0.65 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.85 TMCC3transmembrane and coiled-coil domain family 3 (ENSG00000057704), score: 0.55 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.84 TMEM175transmembrane protein 175 (ENSG00000127419), score: 0.66 TMEM179transmembrane protein 179 (ENSG00000189203), score: 0.59 TMEM184Btransmembrane protein 184B (ENSG00000198792), score: 0.56 TMEM199transmembrane protein 199 (ENSG00000244045), score: -0.47 TMTC2transmembrane and tetratricopeptide repeat containing 2 (ENSG00000179104), score: 0.64 TOMM70Atranslocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) (ENSG00000154174), score: -0.5 TOP1topoisomerase (DNA) I (ENSG00000198900), score: -0.46 TPPPtubulin polymerization promoting protein (ENSG00000171368), score: 0.56 TRDMT1tRNA aspartic acid methyltransferase 1 (ENSG00000107614), score: -0.46 TRIP4thyroid hormone receptor interactor 4 (ENSG00000103671), score: -0.46 TRMT61AtRNA methyltransferase 61 homolog A (S. cerevisiae) (ENSG00000166166), score: 0.56 TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: -0.56 TSPAN15tetraspanin 15 (ENSG00000099282), score: 0.62 TTC15tetratricopeptide repeat domain 15 (ENSG00000171853), score: 0.52 TTC39Btetratricopeptide repeat domain 39B (ENSG00000155158), score: -0.45 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: -0.49 UGT8UDP glycosyltransferase 8 (ENSG00000174607), score: 0.52 UNC5Bunc-5 homolog B (C. elegans) (ENSG00000107731), score: 0.54 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: -0.46 USP3ubiquitin specific peptidase 3 (ENSG00000140455), score: -0.47 USP54ubiquitin specific peptidase 54 (ENSG00000166348), score: 0.61 UTP11LUTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) (ENSG00000183520), score: -0.56 VCAM1vascular cell adhesion molecule 1 (ENSG00000162692), score: -0.44 WBP1WW domain binding protein 1 (ENSG00000115274), score: 0.53 WDR36WD repeat domain 36 (ENSG00000134987), score: -0.54 WDR43WD repeat domain 43 (ENSG00000163811), score: -0.5 WEE1WEE1 homolog (S. pombe) (ENSG00000166483), score: -0.56 WIPF1WAS/WASL interacting protein family, member 1 (ENSG00000115935), score: 0.55 WRBtryptophan rich basic protein (ENSG00000182093), score: 0.53 XRCC4X-ray repair complementing defective repair in Chinese hamster cells 4 (ENSG00000152422), score: -0.45 XRN15'-3' exoribonuclease 1 (ENSG00000114127), score: -0.57 YME1L1YME1-like 1 (S. cerevisiae) (ENSG00000136758), score: -0.56 ZBTB17zinc finger and BTB domain containing 17 (ENSG00000116809), score: 0.6 ZCCHC8zinc finger, CCHC domain containing 8 (ENSG00000033030), score: -0.52 ZDHHC22zinc finger, DHHC-type containing 22 (ENSG00000177108), score: 0.52 ZEB2zinc finger E-box binding homeobox 2 (ENSG00000169554), score: 0.65 ZFAND6zinc finger, AN1-type domain 6 (ENSG00000086666), score: -0.56 ZFYVE20zinc finger, FYVE domain containing 20 (ENSG00000131381), score: 0.54 ZNF276zinc finger protein 276 (ENSG00000158805), score: 0.52 ZNF414zinc finger protein 414 (ENSG00000133250), score: 0.49 ZNF536zinc finger protein 536 (ENSG00000198597), score: 0.64
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |