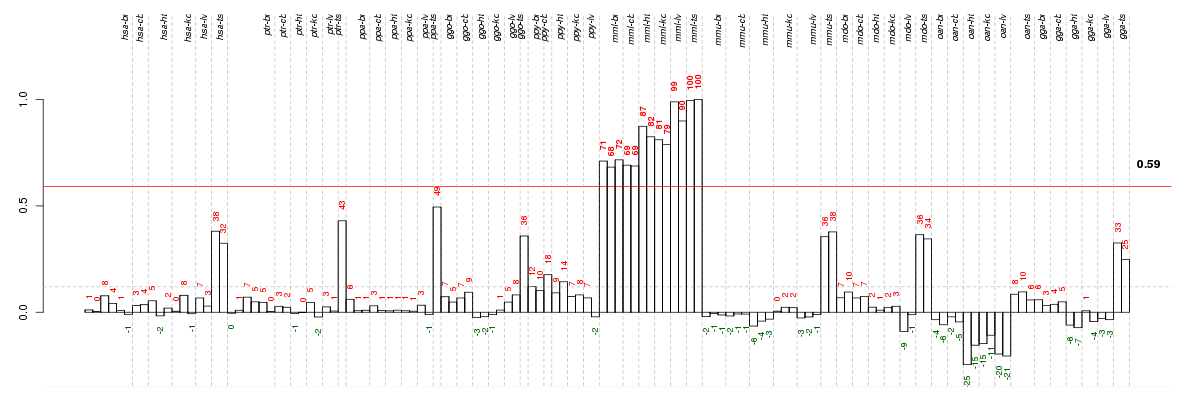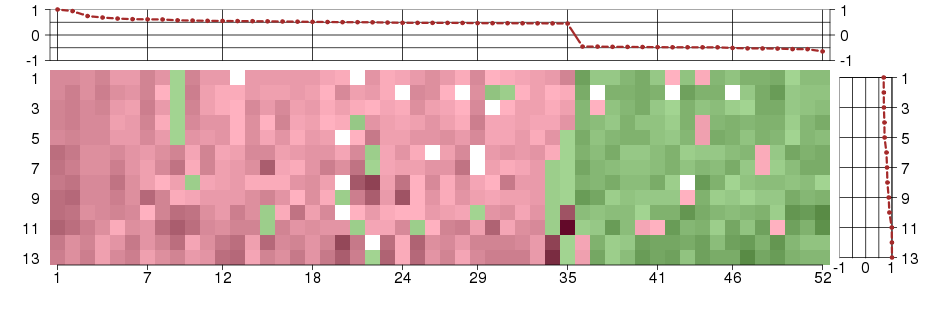

Under-expression is coded with green,
over-expression with red color.



ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.57 ANKLE2ankyrin repeat and LEM domain containing 2 (ENSG00000176915), score: 0.49 ANP32Eacidic (leucine-rich) nuclear phosphoprotein 32 family, member E (ENSG00000143401), score: -0.51 ANXA10annexin A10 (ENSG00000109511), score: 0.58 C12orf26chromosome 12 open reading frame 26 (ENSG00000127720), score: -0.48 C20orf7chromosome 20 open reading frame 7 (ENSG00000101247), score: -0.64 C2CD3C2 calcium-dependent domain containing 3 (ENSG00000168014), score: 0.48 C9orf102chromosome 9 open reading frame 102 (ENSG00000182150), score: 0.52 C9orf16chromosome 9 open reading frame 16 (ENSG00000171159), score: -0.48 CCNJcyclin J (ENSG00000107443), score: 0.5 CHEK2CHK2 checkpoint homolog (S. pombe) (ENSG00000183765), score: -0.47 CHIC2cysteine-rich hydrophobic domain 2 (ENSG00000109220), score: 0.64 CHRNA1cholinergic receptor, nicotinic, alpha 1 (muscle) (ENSG00000138435), score: 0.5 CLCA2chloride channel accessory 2 (ENSG00000137975), score: 0.45 DCAF5DDB1 and CUL4 associated factor 5 (ENSG00000139990), score: 0.55 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (ENSG00000168259), score: -0.55 EIF3Ieukaryotic translation initiation factor 3, subunit I (ENSG00000084623), score: -0.53 EME1essential meiotic endonuclease 1 homolog 1 (S. pombe) (ENSG00000154920), score: 0.56 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: 0.52 FAM184Bfamily with sequence similarity 184, member B (ENSG00000047662), score: 0.69 FBF1Fas (TNFRSF6) binding factor 1 (ENSG00000188878), score: 0.46 GFRALGDNF family receptor alpha like (ENSG00000187871), score: 0.54 HAS3hyaluronan synthase 3 (ENSG00000103044), score: 0.54 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (ENSG00000169087), score: -0.46 MAKmale germ cell-associated kinase (ENSG00000111837), score: 0.62 MASTLmicrotubule associated serine/threonine kinase-like (ENSG00000120539), score: 0.46 MGAT4Amannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A (ENSG00000071073), score: 0.46 MON1AMON1 homolog A (yeast) (ENSG00000164077), score: 0.46 MST1Rmacrophage stimulating 1 receptor (c-met-related tyrosine kinase) (ENSG00000164078), score: 0.61 NRF1nuclear respiratory factor 1 (ENSG00000106459), score: -0.48 NSMCE4Anon-SMC element 4 homolog A (S. cerevisiae) (ENSG00000107672), score: -0.53 PDCD2Lprogrammed cell death 2-like (ENSG00000126249), score: -0.45 PDFpeptide deformylase (mitochondrial) (ENSG00000213380), score: 0.48 PEX3peroxisomal biogenesis factor 3 (ENSG00000034693), score: 0.51 PLIN1perilipin 1 (ENSG00000166819), score: 0.48 PRDX1peroxiredoxin 1 (ENSG00000117450), score: -0.53 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (ENSG00000159377), score: -0.47 RSBN1Lround spermatid basic protein 1-like (ENSG00000187257), score: -0.46 SCLT1sodium channel and clathrin linker 1 (ENSG00000151466), score: 0.94 SEPT12septin 12 (ENSG00000140623), score: 0.74 SERINC4serine incorporator 4 (ENSG00000184716), score: 1 SETDB2SET domain, bifurcated 2 (ENSG00000136169), score: 0.5 SMAD9SMAD family member 9 (ENSG00000120693), score: 0.46 SPOPLspeckle-type POZ protein-like (ENSG00000144228), score: -0.48 TMPRSS9transmembrane protease, serine 9 (ENSG00000178297), score: 0.53 TNNtenascin N (ENSG00000120332), score: 0.61 TRIM35tripartite motif-containing 35 (ENSG00000104228), score: -0.48 TUBD1tubulin, delta 1 (ENSG00000108423), score: 0.48 VPS33Avacuolar protein sorting 33 homolog A (S. cerevisiae) (ENSG00000139719), score: 0.55 WARS2tryptophanyl tRNA synthetase 2, mitochondrial (ENSG00000116874), score: 0.47 ZNHIT3zinc finger, HIT type 3 (ENSG00000108278), score: -0.56
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_br_m1_ca1 | mml | br | m | 1 |
| mml_cb_f_ca1 | mml | cb | f | _ |
| mml_cb_m_ca1 | mml | cb | m | _ |
| mml_br_m2_ca1 | mml | br | m | 2 |
| mml_br_f_ca1 | mml | br | f | _ |
| mml_kd_f_ca1 | mml | kd | f | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |
| mml_ht_f_ca1 | mml | ht | f | _ |
| mml_ht_m_ca1 | mml | ht | m | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |
| mml_ts_m1_ca1 | mml | ts | m | 1 |
| mml_ts_m2_ca1 | mml | ts | m | 2 |