

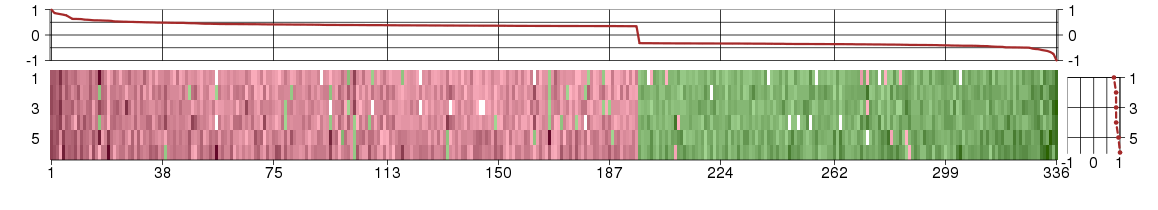

Under-expression is coded with green,
over-expression with red color.



AAGABalpha- and gamma-adaptin binding protein (ENSG00000103591), score: -0.59 ABCC1ATP-binding cassette, sub-family C (CFTR/MRP), member 1 (ENSG00000103222), score: 0.49 ABL1c-abl oncogene 1, non-receptor tyrosine kinase (ENSG00000097007), score: 0.35 ABLIM1actin binding LIM protein 1 (ENSG00000099204), score: 0.38 ABTB1ankyrin repeat and BTB (POZ) domain containing 1 (ENSG00000114626), score: 0.48 ADAM19ADAM metallopeptidase domain 19 (ENSG00000135074), score: 0.41 ADAMTS14ADAM metallopeptidase with thrombospondin type 1 motif, 14 (ENSG00000138316), score: 0.52 AGFG1ArfGAP with FG repeats 1 (ENSG00000173744), score: -0.34 AHCTF1AT hook containing transcription factor 1 (ENSG00000153207), score: -0.63 AHSA2AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) (ENSG00000173209), score: -0.4 ANKRD28ankyrin repeat domain 28 (ENSG00000206560), score: -0.36 ANKRD54ankyrin repeat domain 54 (ENSG00000100124), score: -0.46 ANKS6ankyrin repeat and sterile alpha motif domain containing 6 (ENSG00000165138), score: 0.39 AP4B1adaptor-related protein complex 4, beta 1 subunit (ENSG00000134262), score: 0.46 APCDD1adenomatosis polyposis coli down-regulated 1 (ENSG00000154856), score: 0.44 ARCN1archain 1 (ENSG00000095139), score: -0.48 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (ENSG00000101199), score: 0.56 ARFRP1ADP-ribosylation factor related protein 1 (ENSG00000101246), score: 0.35 ARHGEF10Rho guanine nucleotide exchange factor (GEF) 10 (ENSG00000104728), score: 0.35 ATG10ATG10 autophagy related 10 homolog (S. cerevisiae) (ENSG00000152348), score: -0.34 ATL2atlastin GTPase 2 (ENSG00000119787), score: -0.49 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: 0.42 AXIN2axin 2 (ENSG00000168646), score: 0.35 AZI25-azacytidine induced 2 (ENSG00000163512), score: -0.35 BACH1BTB and CNC homology 1, basic leucine zipper transcription factor 1 (ENSG00000156273), score: -0.35 BARX1BARX homeobox 1 (ENSG00000131668), score: 0.41 BEST1bestrophin 1 (ENSG00000167995), score: 0.47 BMP7bone morphogenetic protein 7 (ENSG00000101144), score: 0.37 BUB1Bbudding uninhibited by benzimidazoles 1 homolog beta (yeast) (ENSG00000156970), score: 0.57 C10orf46chromosome 10 open reading frame 46 (ENSG00000151893), score: -0.33 C10orf76chromosome 10 open reading frame 76 (ENSG00000120029), score: 0.44 C12orf29chromosome 12 open reading frame 29 (ENSG00000133641), score: -0.33 C14orf143chromosome 14 open reading frame 143 (ENSG00000140025), score: -0.33 C14orf159chromosome 14 open reading frame 159 (ENSG00000133943), score: 0.37 C15orf17chromosome 15 open reading frame 17 (ENSG00000178761), score: 0.36 C16orf57chromosome 16 open reading frame 57 (ENSG00000103005), score: 0.36 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: 0.38 C1orf151chromosome 1 open reading frame 151 (ENSG00000173436), score: 0.37 C3orf18chromosome 3 open reading frame 18 (ENSG00000088543), score: 0.44 C6orf115chromosome 6 open reading frame 115 (ENSG00000146386), score: -0.37 C7orf23chromosome 7 open reading frame 23 (ENSG00000135185), score: -0.38 C8orf45chromosome 8 open reading frame 45 (ENSG00000178460), score: 0.99 C8orf79chromosome 8 open reading frame 79 (ENSG00000170941), score: 0.37 CACNA2D4calcium channel, voltage-dependent, alpha 2/delta subunit 4 (ENSG00000151062), score: 0.84 CBFBcore-binding factor, beta subunit (ENSG00000067955), score: 0.41 CBX7chromobox homolog 7 (ENSG00000100307), score: 0.42 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: 0.43 CCDC51coiled-coil domain containing 51 (ENSG00000164051), score: 0.52 CCDC6coiled-coil domain containing 6 (ENSG00000108091), score: -0.38 CCDC90Bcoiled-coil domain containing 90B (ENSG00000137500), score: -0.49 CCNHcyclin H (ENSG00000134480), score: -0.36 CCNJLcyclin J-like (ENSG00000135083), score: 0.39 CCT2chaperonin containing TCP1, subunit 2 (beta) (ENSG00000166226), score: -0.39 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: -0.34 CHMP2Bchromatin modifying protein 2B (ENSG00000083937), score: -0.34 CHRNA10cholinergic receptor, nicotinic, alpha 10 (ENSG00000129749), score: 0.38 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.37 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: -0.34 CLINT1clathrin interactor 1 (ENSG00000113282), score: -0.33 CLN5ceroid-lipofuscinosis, neuronal 5 (ENSG00000102805), score: -0.37 COL23A1collagen, type XXIII, alpha 1 (ENSG00000050767), score: 0.35 COL7A1collagen, type VII, alpha 1 (ENSG00000114270), score: 0.37 COL8A1collagen, type VIII, alpha 1 (ENSG00000144810), score: 0.41 COLQcollagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase (ENSG00000206561), score: 0.6 COX18COX18 cytochrome c oxidase assembly homolog (S. cerevisiae) (ENSG00000163626), score: -0.41 CPSF4Lcleavage and polyadenylation specific factor 4-like (ENSG00000187959), score: 0.7 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: -0.43 CTNNBIP1catenin, beta interacting protein 1 (ENSG00000178585), score: 0.4 CUEDC2CUE domain containing 2 (ENSG00000107874), score: 0.36 CUL1cullin 1 (ENSG00000055130), score: -1 CUL3cullin 3 (ENSG00000036257), score: -0.42 CWF19L1CWF19-like 1, cell cycle control (S. pombe) (ENSG00000095485), score: -0.37 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: -0.4 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: -0.56 DCTdopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) (ENSG00000080166), score: 0.51 DDX51DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 (ENSG00000185163), score: 0.42 DDX55DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 (ENSG00000111364), score: 0.46 DEPDC7DEP domain containing 7 (ENSG00000121690), score: -0.36 DFNA5deafness, autosomal dominant 5 (ENSG00000105928), score: 0.35 DHRS11dehydrogenase/reductase (SDR family) member 11 (ENSG00000108272), score: 0.36 DMAP1DNA methyltransferase 1 associated protein 1 (ENSG00000178028), score: 0.4 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: -0.49 DNAJC3DnaJ (Hsp40) homolog, subfamily C, member 3 (ENSG00000102580), score: -0.32 DRAM2DNA-damage regulated autophagy modulator 2 (ENSG00000156171), score: -0.32 DUSP26dual specificity phosphatase 26 (putative) (ENSG00000133878), score: 0.38 DVL3dishevelled, dsh homolog 3 (Drosophila) (ENSG00000161202), score: 0.63 DYNC2LI1dynein, cytoplasmic 2, light intermediate chain 1 (ENSG00000138036), score: -0.42 E2F5E2F transcription factor 5, p130-binding (ENSG00000133740), score: -0.42 E4F1E4F transcription factor 1 (ENSG00000167967), score: 0.42 EEDembryonic ectoderm development (ENSG00000074266), score: 0.36 EIF2B4eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa (ENSG00000115211), score: 0.37 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.46 ENTPD7ectonucleoside triphosphate diphosphohydrolase 7 (ENSG00000198018), score: -0.42 EPS15L1epidermal growth factor receptor pathway substrate 15-like 1 (ENSG00000127527), score: 0.4 EPT1ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) (ENSG00000138018), score: -0.49 ERAL1Era G-protein-like 1 (E. coli) (ENSG00000132591), score: 0.41 ESYT2extended synaptotagmin-like protein 2 (ENSG00000117868), score: 0.36 ETAA1Ewing tumor-associated antigen 1 (ENSG00000143971), score: -0.49 EXOC3exocyst complex component 3 (ENSG00000180104), score: 0.36 EXOSC8exosome component 8 (ENSG00000120699), score: -0.45 FAM134Cfamily with sequence similarity 134, member C (ENSG00000141699), score: -0.39 FAM151Bfamily with sequence similarity 151, member B (ENSG00000152380), score: -0.42 FAM168Bfamily with sequence similarity 168, member B (ENSG00000152102), score: 0.36 FAM189A2family with sequence similarity 189, member A2 (ENSG00000135063), score: 0.35 FAM59Bfamily with sequence similarity 59, member B (ENSG00000157833), score: 0.62 FAM69Cfamily with sequence similarity 69, member C (ENSG00000187773), score: 0.49 FAM8A1family with sequence similarity 8, member A1 (ENSG00000137414), score: -0.34 FBRSL1fibrosin-like 1 (ENSG00000112787), score: 0.41 FBXW4F-box and WD repeat domain containing 4 (ENSG00000107829), score: 0.36 FER1L5fer-1-like 5 (C. elegans) (ENSG00000214272), score: -0.33 FKBP10FK506 binding protein 10, 65 kDa (ENSG00000141756), score: 0.37 FKTNfukutin (ENSG00000106692), score: -0.33 FNDC3Afibronectin type III domain containing 3A (ENSG00000102531), score: -0.36 FRMD4AFERM domain containing 4A (ENSG00000151474), score: 0.4 FRZBfrizzled-related protein (ENSG00000162998), score: 0.41 FTSJD2FtsJ methyltransferase domain containing 2 (ENSG00000137200), score: 0.37 GALK2galactokinase 2 (ENSG00000156958), score: -0.33 GEMIN6gem (nuclear organelle) associated protein 6 (ENSG00000152147), score: -0.34 GFPT2glutamine-fructose-6-phosphate transaminase 2 (ENSG00000131459), score: 0.44 GLG1golgi glycoprotein 1 (ENSG00000090863), score: 0.43 GLIPR2GLI pathogenesis-related 2 (ENSG00000122694), score: 0.36 GORASP1golgi reassembly stacking protein 1, 65kDa (ENSG00000114745), score: 0.41 GPATCH1G patch domain containing 1 (ENSG00000076650), score: 0.37 GPC1glypican 1 (ENSG00000063660), score: 0.48 GRAMD1CGRAM domain containing 1C (ENSG00000178075), score: -0.39 GUCA1Aguanylate cyclase activator 1A (retina) (ENSG00000048545), score: 0.39 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: -0.39 H2AFYH2A histone family, member Y (ENSG00000113648), score: 0.39 HDDC2HD domain containing 2 (ENSG00000111906), score: 0.48 HERPUD2HERPUD family member 2 (ENSG00000122557), score: -0.32 HESX1HESX homeobox 1 (ENSG00000163666), score: 0.35 HIAT1hippocampus abundant transcript 1 (ENSG00000156875), score: -0.33 HKDC1hexokinase domain containing 1 (ENSG00000156510), score: 0.86 HM13histocompatibility (minor) 13 (ENSG00000101294), score: -0.41 HMHA1histocompatibility (minor) HA-1 (ENSG00000180448), score: 0.38 HSF4heat shock transcription factor 4 (ENSG00000102878), score: 0.36 HSPA13heat shock protein 70kDa family, member 13 (ENSG00000155304), score: -0.42 HTRA1HtrA serine peptidase 1 (ENSG00000166033), score: 0.35 IL17Binterleukin 17B (ENSG00000127743), score: 0.56 IL17RAinterleukin 17 receptor A (ENSG00000177663), score: 0.59 IL20RAinterleukin 20 receptor, alpha (ENSG00000016402), score: 0.42 INHAinhibin, alpha (ENSG00000123999), score: 0.51 IP6K3inositol hexakisphosphate kinase 3 (ENSG00000161896), score: 0.51 IQCEIQ motif containing E (ENSG00000106012), score: 0.36 ITGA11integrin, alpha 11 (ENSG00000137809), score: 0.77 JAG2jagged 2 (ENSG00000184916), score: 0.39 KBTBD5kelch repeat and BTB (POZ) domain containing 5 (ENSG00000157119), score: 0.4 KCTD20potassium channel tetramerisation domain containing 20 (ENSG00000112078), score: -0.42 KDM3Blysine (K)-specific demethylase 3B (ENSG00000120733), score: 0.35 KIAA0020KIAA0020 (ENSG00000080608), score: -0.38 KIAA1033KIAA1033 (ENSG00000136051), score: -0.35 KIAA1712KIAA1712 (ENSG00000164118), score: -0.38 KIF15kinesin family member 15 (ENSG00000163808), score: 0.57 KLHDC10kelch domain containing 10 (ENSG00000128607), score: -0.33 LDLRAP1low density lipoprotein receptor adaptor protein 1 (ENSG00000157978), score: 0.35 LOC100131509similar to inward rectifying K+ channel negative regulator Kir2.2v (ENSG00000184185), score: 0.35 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: 0.51 LOC100289938similar to cytoplasmic tRNA 2-thiolation protein 2 (ENSG00000174177), score: 0.48 LOC100291671similar to SH3-binding domain and glutamic acid-rich protein (ENSG00000185437), score: 0.35 LPCAT1lysophosphatidylcholine acyltransferase 1 (ENSG00000153395), score: 0.43 LPGAT1lysophosphatidylglycerol acyltransferase 1 (ENSG00000123684), score: -0.32 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (ENSG00000198799), score: 0.42 LRP2BPLRP2 binding protein (ENSG00000109771), score: 0.38 LRRC14Bleucine rich repeat containing 14B (ENSG00000185028), score: 0.41 LRRC33leucine rich repeat containing 33 (ENSG00000174004), score: 0.35 LRRC40leucine rich repeat containing 40 (ENSG00000066557), score: -0.34 LUC7L2LUC7-like 2 (S. cerevisiae) (ENSG00000146963), score: 0.37 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: 0.45 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (ENSG00000198517), score: 0.35 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: -0.32 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: 0.55 MAPK12mitogen-activated protein kinase 12 (ENSG00000188130), score: 0.48 MARCH2membrane-associated ring finger (C3HC4) 2 (ENSG00000099785), score: 0.35 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: -0.33 MCF2LMCF.2 cell line derived transforming sequence-like (ENSG00000126217), score: 0.36 MCM3APminichromosome maintenance complex component 3 associated protein (ENSG00000160294), score: 0.36 MED22mediator complex subunit 22 (ENSG00000148297), score: 0.49 MED7mediator complex subunit 7 (ENSG00000155868), score: -0.33 MEGF6multiple EGF-like-domains 6 (ENSG00000162591), score: 0.35 MFSD2Bmajor facilitator superfamily domain containing 2B (ENSG00000205639), score: 0.82 MGEA5meningioma expressed antigen 5 (hyaluronidase) (ENSG00000198408), score: 0.48 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: -0.4 MKL1megakaryoblastic leukemia (translocation) 1 (ENSG00000196588), score: 0.35 MMAAmethylmalonic aciduria (cobalamin deficiency) cblA type (ENSG00000151611), score: -0.41 MORN4MORN repeat containing 4 (ENSG00000171160), score: 0.39 MPHOSPH8M-phase phosphoprotein 8 (ENSG00000196199), score: 0.5 MPP6membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (ENSG00000105926), score: -0.33 MRPS23mitochondrial ribosomal protein S23 (ENSG00000181610), score: 0.39 MSX1msh homeobox 1 (ENSG00000163132), score: 0.42 MSX2msh homeobox 2 (ENSG00000120149), score: 0.42 MYCBPAPMYCBP associated protein (ENSG00000136449), score: 0.37 MYO18Amyosin XVIIIA (ENSG00000196535), score: 0.36 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.43 NCBP1nuclear cap binding protein subunit 1, 80kDa (ENSG00000136937), score: -0.33 ND3NADH dehydrogenase, subunit 3 (complex I) (ENSG00000198840), score: 0.37 ND4NADH dehydrogenase, subunit 4 (complex I) (ENSG00000198886), score: 0.37 NDFIP1Nedd4 family interacting protein 1 (ENSG00000131507), score: -0.32 NDUFB2NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa (ENSG00000090266), score: -0.39 NME7non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000143156), score: -0.55 NOD1nucleotide-binding oligomerization domain containing 1 (ENSG00000106100), score: 0.35 NOL6nucleolar protein family 6 (RNA-associated) (ENSG00000165271), score: 0.5 NPATnuclear protein, ataxia-telangiectasia locus (ENSG00000149308), score: -0.33 NRBP2nuclear receptor binding protein 2 (ENSG00000185189), score: 0.35 NRF1nuclear respiratory factor 1 (ENSG00000106459), score: 0.41 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: 0.38 NUMA1nuclear mitotic apparatus protein 1 (ENSG00000137497), score: 0.4 NUP155nucleoporin 155kDa (ENSG00000113569), score: -0.33 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (ENSG00000173559), score: -0.37 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: 0.37 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: 0.37 PANK3pantothenate kinase 3 (ENSG00000120137), score: -0.34 PARP1poly (ADP-ribose) polymerase 1 (ENSG00000143799), score: 0.35 PCGF2polycomb group ring finger 2 (ENSG00000056661), score: 0.43 PCID2PCI domain containing 2 (ENSG00000126226), score: 0.5 PCYOX1prenylcysteine oxidase 1 (ENSG00000116005), score: -0.34 PDAP1PDGFA associated protein 1 (ENSG00000106244), score: 0.63 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.6 PDE9Aphosphodiesterase 9A (ENSG00000160191), score: 0.38 PEX3peroxisomal biogenesis factor 3 (ENSG00000034693), score: -0.39 PHF10PHD finger protein 10 (ENSG00000130024), score: -0.46 PHTF2putative homeodomain transcription factor 2 (ENSG00000006576), score: -0.37 PIGPphosphatidylinositol glycan anchor biosynthesis, class P (ENSG00000185808), score: -0.33 PITPNC1phosphatidylinositol transfer protein, cytoplasmic 1 (ENSG00000154217), score: 0.39 PKNOX1PBX/knotted 1 homeobox 1 (ENSG00000160199), score: -0.33 PKP1plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) (ENSG00000081277), score: 0.57 PLAGL1pleiomorphic adenoma gene-like 1 (ENSG00000118495), score: 0.38 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (ENSG00000161980), score: 0.37 PPIL3peptidylprolyl isomerase (cyclophilin)-like 3 (ENSG00000240344), score: -0.67 PRELPproline/arginine-rich end leucine-rich repeat protein (ENSG00000188783), score: 0.43 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (ENSG00000101161), score: 0.43 PSMA3proteasome (prosome, macropain) subunit, alpha type, 3 (ENSG00000100567), score: -0.53 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (ENSG00000159377), score: 0.35 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (ENSG00000197170), score: -0.37 PSTKphosphoseryl-tRNA kinase (ENSG00000179988), score: -0.35 PTCD1pentatricopeptide repeat domain 1 (ENSG00000106246), score: 0.8 PUS3pseudouridylate synthase 3 (ENSG00000110060), score: -0.33 PUS7pseudouridylate synthase 7 homolog (S. cerevisiae) (ENSG00000091127), score: -0.43 PVRL3poliovirus receptor-related 3 (ENSG00000177707), score: -0.44 QSER1glutamine and serine rich 1 (ENSG00000060749), score: -0.37 RALAv-ral simian leukemia viral oncogene homolog A (ras related) (ENSG00000006451), score: -0.74 RBM45RNA binding motif protein 45 (ENSG00000155636), score: -0.34 RBM6RNA binding motif protein 6 (ENSG00000004534), score: 0.39 RBM7RNA binding motif protein 7 (ENSG00000076053), score: -0.36 RECQL4RecQ protein-like 4 (ENSG00000160957), score: 0.63 RGS11regulator of G-protein signaling 11 (ENSG00000076344), score: 0.4 RHBDL3rhomboid, veinlet-like 3 (Drosophila) (ENSG00000141314), score: 0.39 RMND1required for meiotic nuclear division 1 homolog (S. cerevisiae) (ENSG00000155906), score: -0.46 RNF103ring finger protein 103 (ENSG00000239305), score: 0.38 RNF139ring finger protein 139 (ENSG00000170881), score: -0.33 RNF144Aring finger protein 144A (ENSG00000151692), score: 0.36 RNMTL1RNA methyltransferase like 1 (ENSG00000171861), score: 0.35 ROBO3roundabout, axon guidance receptor, homolog 3 (Drosophila) (ENSG00000154134), score: 0.53 RPS6KA2ribosomal protein S6 kinase, 90kDa, polypeptide 2 (ENSG00000071242), score: 0.4 RRHretinal pigment epithelium-derived rhodopsin homolog (ENSG00000180245), score: 0.48 RUNX1T1runt-related transcription factor 1; translocated to, 1 (cyclin D-related) (ENSG00000079102), score: 0.35 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: 0.41 SEC23IPSEC23 interacting protein (ENSG00000107651), score: -0.33 SEC61A1Sec61 alpha 1 subunit (S. cerevisiae) (ENSG00000058262), score: -0.34 SEC63SEC63 homolog (S. cerevisiae) (ENSG00000025796), score: -0.35 SEPSECSSep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (ENSG00000109618), score: -0.34 SFRS18splicing factor, arginine/serine-rich 18 (ENSG00000132424), score: 0.43 SGPL1sphingosine-1-phosphate lyase 1 (ENSG00000166224), score: -0.36 SGPP1sphingosine-1-phosphate phosphatase 1 (ENSG00000126821), score: -0.38 SH2D3CSH2 domain containing 3C (ENSG00000095370), score: 0.42 SIKE1suppressor of IKBKE 1 (ENSG00000052723), score: -0.33 SKILSKI-like oncogene (ENSG00000136603), score: -0.38 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: -0.32 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: -0.48 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: -0.38 SLC35D1solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 (ENSG00000116704), score: -0.37 SLCO4A1solute carrier organic anion transporter family, member 4A1 (ENSG00000101187), score: 0.37 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: -0.35 SNED1sushi, nidogen and EGF-like domains 1 (ENSG00000162804), score: 0.39 SNX14sorting nexin 14 (ENSG00000135317), score: -0.34 SNX33sorting nexin 33 (ENSG00000173548), score: 0.38 SPTBN5spectrin, beta, non-erythrocytic 5 (ENSG00000137877), score: 0.36 SSR1signal sequence receptor, alpha (ENSG00000124783), score: -0.37 SSRP1structure specific recognition protein 1 (ENSG00000149136), score: 0.35 STARD3StAR-related lipid transfer (START) domain containing 3 (ENSG00000131748), score: 0.38 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: -0.36 SYCP2Lsynaptonemal complex protein 2-like (ENSG00000153157), score: 0.37 SYNMsynemin, intermediate filament protein (ENSG00000182253), score: 0.44 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (ENSG00000172660), score: 0.58 TAF1DTATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa (ENSG00000166012), score: 0.37 TARSL2threonyl-tRNA synthetase-like 2 (ENSG00000185418), score: 0.37 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (ENSG00000133138), score: -0.36 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (ENSG00000100726), score: 0.53 TERF2telomeric repeat binding factor 2 (ENSG00000132604), score: 0.35 TIMM22translocase of inner mitochondrial membrane 22 homolog (yeast) (ENSG00000177370), score: 0.39 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: 0.53 TLK1tousled-like kinase 1 (ENSG00000198586), score: -0.37 TM9SF2transmembrane 9 superfamily member 2 (ENSG00000125304), score: -0.36 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.35 TMCO1transmembrane and coiled-coil domains 1 (ENSG00000143183), score: -0.36 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: -0.47 TMEM135transmembrane protein 135 (ENSG00000166575), score: -0.33 TMEM33transmembrane protein 33 (ENSG00000109133), score: -0.35 TMEM41Btransmembrane protein 41B (ENSG00000166471), score: -0.41 TOM1L2target of myb1-like 2 (chicken) (ENSG00000175662), score: 0.37 TOMM70Atranslocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) (ENSG00000154174), score: -0.4 TOX3TOX high mobility group box family member 3 (ENSG00000103460), score: -0.33 TPOthyroid peroxidase (ENSG00000115705), score: 0.35 TRIM7tripartite motif-containing 7 (ENSG00000146054), score: 0.35 TSEN2tRNA splicing endonuclease 2 homolog (S. cerevisiae) (ENSG00000154743), score: 0.41 TSHRthyroid stimulating hormone receptor (ENSG00000165409), score: 0.45 TSPAN15tetraspanin 15 (ENSG00000099282), score: 0.39 TSPAN18tetraspanin 18 (ENSG00000157570), score: 0.39 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: 0.37 TTC1tetratricopeptide repeat domain 1 (ENSG00000113312), score: 0.35 TTC27tetratricopeptide repeat domain 27 (ENSG00000018699), score: -0.33 TTC33tetratricopeptide repeat domain 33 (ENSG00000113638), score: -0.36 UBE2Wubiquitin-conjugating enzyme E2W (putative) (ENSG00000104343), score: -0.34 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: 0.4 UBL3ubiquitin-like 3 (ENSG00000122042), score: -0.39 UBR7ubiquitin protein ligase E3 component n-recognin 7 (putative) (ENSG00000012963), score: -0.44 UBXN4UBX domain protein 4 (ENSG00000144224), score: -0.49 UHRF1BP1LUHRF1 binding protein 1-like (ENSG00000111647), score: -0.35 UPP1uridine phosphorylase 1 (ENSG00000183696), score: 0.39 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: -0.36 USP14ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) (ENSG00000101557), score: -0.35 USP45ubiquitin specific peptidase 45 (ENSG00000123552), score: -0.33 VITvitrin (ENSG00000205221), score: 0.49 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: -0.35 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: -0.61 WDR37WD repeat domain 37 (ENSG00000047056), score: 0.38 WDR60WD repeat domain 60 (ENSG00000126870), score: 0.38 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: -0.5 XRCC3X-ray repair complementing defective repair in Chinese hamster cells 3 (ENSG00000126215), score: 0.48 XRN15'-3' exoribonuclease 1 (ENSG00000114127), score: -0.37 XRN25'-3' exoribonuclease 2 (ENSG00000088930), score: -0.35 ZC4H2zinc finger, C4H2 domain containing (ENSG00000126970), score: 0.38 ZDHHC12zinc finger, DHHC-type containing 12 (ENSG00000160446), score: -0.34 ZFYVE21zinc finger, FYVE domain containing 21 (ENSG00000100711), score: 0.43 ZNF800zinc finger protein 800 (ENSG00000048405), score: -0.35 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: 0.39 ZW10ZW10, kinetochore associated, homolog (Drosophila) (ENSG00000086827), score: -0.39
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_br_f_ca1 | ggo | br | f | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ggo_br_m_ca1 | ggo | br | m | _ |
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| ggo_ht_m_ca1 | ggo | ht | m | _ |
| ggo_ht_f_ca1 | ggo | ht | f | _ |
