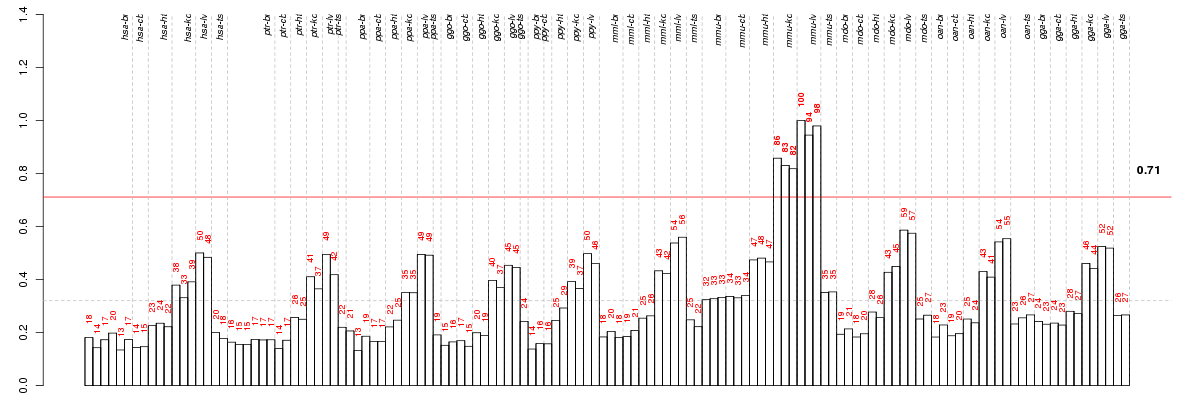

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
oxidation reduction
The process of removal or addition of one or more electrons with or without the concomitant removal or addition of a proton or protons.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
coenzyme metabolic process
The chemical reactions and pathways involving coenzymes, any of various nonprotein organic cofactors that are required, in addition to an enzyme and a substrate, for an enzymatic reaction to proceed.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
monocarboxylic acid metabolic process
The chemical reactions and pathways involving monocarboxylic acids, any organic acid containing one carboxyl (COOH) group or anion (COO-).
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
cofactor metabolic process
The chemical reactions and pathways involving a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
peroxisome
A small, membrane-bounded organelle that uses dioxygen (O2) to oxidize organic molecules; contains some enzymes that produce and others that degrade hydrogen peroxide (H2O2).
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
organelle membrane
The lipid bilayer surrounding an organelle.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.

nucleotide binding
Interacting selectively and non-covalently with a nucleotide, any compound consisting of a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the ribose or deoxyribose moiety.
nucleoside binding
Interacting selectively and non-covalently with a nucleoside, a compound consisting of a purine or pyrimidine nitrogenous base linked either to ribose or deoxyribose.
purine nucleoside binding
Interacting selectively and non-covalently with a purine nucleoside, a compound consisting of a purine base linked either to ribose or deoxyribose.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
monooxygenase activity
Catalysis of the incorporation of one atom from molecular oxygen into a compound and the reduction of the other atom of oxygen to water.
hydrolase activity, hydrolyzing O-glycosyl compounds
Catalysis of the hydrolysis of any O-glycosyl bond.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
mannosidase activity
Catalysis of the hydrolysis of mannosyl compounds, substances containing a group derived from a cyclic form of mannose or a mannose derivative.
mannosyl-oligosaccharide mannosidase activity
Catalysis of the hydrolysis of the terminal alpha-D-mannose residues in oligo-mannose oligosaccharides.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on glycosyl bonds
Catalysis of the hydrolysis of any glycosyl bond.
purine nucleotide binding
Interacting selectively and non-covalently with purine nucleotides, any compound consisting of a purine nucleoside esterified with (ortho)phosphate.
vitamin binding
Interacting selectively and non-covalently with a vitamin, one of a number of unrelated organic substances that occur in many foods in small amounts and that are necessary in trace amounts for the normal metabolic functioning of the body.
adenyl nucleotide binding
Interacting selectively and non-covalently with adenyl nucleotides, any compound consisting of adenosine esterified with (ortho)phosphate.
cofactor binding
Interacting selectively and non-covalently with a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
FAD binding
Interacting selectively and non-covalently with FAD, flavin-adenine dinucleotide, the coenzyme or the prosthetic group of various flavoprotein oxidoreductase enzymes.
coenzyme binding
Interacting selectively and non-covalently with a coenzyme, any of various nonprotein organic cofactors that are required, in addition to an enzyme and a substrate, for an enzymatic reaction to proceed.
all
NA
adenyl nucleotide binding
Interacting selectively and non-covalently with adenyl nucleotides, any compound consisting of adenosine esterified with (ortho)phosphate.
FAD binding
Interacting selectively and non-covalently with FAD, flavin-adenine dinucleotide, the coenzyme or the prosthetic group of various flavoprotein oxidoreductase enzymes.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 01100 | 8.727e-04 | 24.91 | 45 | 353 | Metabolic pathways |
| 04146 | 1.743e-03 | 2.117 | 10 | 30 | Peroxisome |
| 00260 | 4.569e-02 | 0.9174 | 5 | 13 | Glycine, serine and threonine metabolism |
A1CFAPOBEC1 complementation factor (ENSG00000148584), score: 0.46 A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.73 ABCC2ATP-binding cassette, sub-family C (CFTR/MRP), member 2 (ENSG00000023839), score: 0.54 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.73 ABHD3abhydrolase domain containing 3 (ENSG00000158201), score: 0.46 ABI2abl-interactor 2 (ENSG00000138443), score: -0.54 ACBD5acyl-CoA binding domain containing 5 (ENSG00000107897), score: 0.49 ACO1aconitase 1, soluble (ENSG00000122729), score: 0.5 ACOT12acyl-CoA thioesterase 12 (ENSG00000172497), score: 0.58 ACSL5acyl-CoA synthetase long-chain family member 5 (ENSG00000197142), score: 0.46 ADAP2ArfGAP with dual PH domains 2 (ENSG00000184060), score: 0.57 AGPHD1aminoglycoside phosphotransferase domain containing 1 (ENSG00000188266), score: 0.55 AIM1absent in melanoma 1 (ENSG00000112297), score: 0.41 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (ENSG00000117448), score: 0.51 ALAS1aminolevulinate, delta-, synthase 1 (ENSG00000023330), score: 0.4 ALDH8A1aldehyde dehydrogenase 8 family, member A1 (ENSG00000118514), score: 0.49 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.41 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.62 ANGEL1angel homolog 1 (Drosophila) (ENSG00000013523), score: -0.49 ANGEL2angel homolog 2 (Drosophila) (ENSG00000174606), score: 0.43 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.46 ANKRD16ankyrin repeat domain 16 (ENSG00000134461), score: 0.41 ANKS4Bankyrin repeat and sterile alpha motif domain containing 4B (ENSG00000175311), score: 0.53 AQP8aquaporin 8 (ENSG00000103375), score: 0.47 ARCN1archain 1 (ENSG00000095139), score: 0.49 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.5 ARHGAP42Rho GTPase activating protein 42 (ENSG00000165895), score: 0.64 ARSBarylsulfatase B (ENSG00000113273), score: 0.52 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.54 ATL2atlastin GTPase 2 (ENSG00000119787), score: 0.42 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.41 ATP6V0A2ATPase, H+ transporting, lysosomal V0 subunit a2 (ENSG00000185344), score: 0.42 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: -0.6 ATXN7ataxin 7 (ENSG00000163635), score: 0.44 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.46 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.84 BBS2Bardet-Biedl syndrome 2 (ENSG00000125124), score: -0.61 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: 0.45 BPHLbiphenyl hydrolase-like (serine hydrolase) (ENSG00000137274), score: 0.49 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.75 BRAPBRCA1 associated protein (ENSG00000089234), score: 0.47 BTBD10BTB (POZ) domain containing 10 (ENSG00000148925), score: -0.47 C11orf54chromosome 11 open reading frame 54 (ENSG00000182919), score: 0.57 C12orf72chromosome 12 open reading frame 72 (ENSG00000139160), score: 0.55 C16orf48chromosome 16 open reading frame 48 (ENSG00000124074), score: -0.48 C19orf12chromosome 19 open reading frame 12 (ENSG00000131943), score: 0.44 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.57 C21orf63chromosome 21 open reading frame 63 (ENSG00000166979), score: -0.48 C2CD2C2 calcium-dependent domain containing 2 (ENSG00000157617), score: 0.52 C5orf33chromosome 5 open reading frame 33 (ENSG00000152620), score: 0.45 C6orf106chromosome 6 open reading frame 106 (ENSG00000196821), score: 0.41 C7orf23chromosome 7 open reading frame 23 (ENSG00000135185), score: 0.7 C9orf150chromosome 9 open reading frame 150 (ENSG00000153714), score: 0.42 C9orf95chromosome 9 open reading frame 95 (ENSG00000106733), score: 0.43 CASP8caspase 8, apoptosis-related cysteine peptidase (ENSG00000064012), score: 0.57 CCBL2cysteine conjugate-beta lyase 2 (ENSG00000137944), score: 0.62 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: 0.46 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.59 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.77 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.71 CGRRF1cell growth regulator with ring finger domain 1 (ENSG00000100532), score: 0.51 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.48 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: 0.43 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.69 CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.42 CIRBPcold inducible RNA binding protein (ENSG00000099622), score: -0.52 CKAP5cytoskeleton associated protein 5 (ENSG00000175216), score: -0.51 CLEC3BC-type lectin domain family 3, member B (ENSG00000163815), score: -0.57 CMTM6CKLF-like MARVEL transmembrane domain containing 6 (ENSG00000091317), score: 0.5 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.9 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.76 COMMD3COMM domain containing 3 (ENSG00000148444), score: 0.45 COPB2coatomer protein complex, subunit beta 2 (beta prime) (ENSG00000184432), score: 0.49 CPOXcoproporphyrinogen oxidase (ENSG00000080819), score: 0.55 CROTcarnitine O-octanoyltransferase (ENSG00000005469), score: 0.69 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.45 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: 0.46 CYBRD1cytochrome b reductase 1 (ENSG00000071967), score: -0.56 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (ENSG00000172817), score: 0.56 DARSaspartyl-tRNA synthetase (ENSG00000115866), score: 0.42 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.46 DCLK3doublecortin-like kinase 3 (ENSG00000163673), score: 0.44 DCLRE1ADNA cross-link repair 1A (ENSG00000198924), score: 0.49 DEPDC6DEP domain containing 6 (ENSG00000155792), score: 0.45 DERL2Der1-like domain family, member 2 (ENSG00000072849), score: 0.45 DHCR2424-dehydrocholesterol reductase (ENSG00000116133), score: 0.41 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: -0.57 DLEC1deleted in lung and esophageal cancer 1 (ENSG00000008226), score: 0.51 DMGDHdimethylglycine dehydrogenase (ENSG00000132837), score: 0.47 DNAJC22DnaJ (Hsp40) homolog, subfamily C, member 22 (ENSG00000178401), score: 0.58 DNAJC3DnaJ (Hsp40) homolog, subfamily C, member 3 (ENSG00000102580), score: 0.59 DNTTdeoxynucleotidyltransferase, terminal (ENSG00000107447), score: 0.52 DPP4dipeptidyl-peptidase 4 (ENSG00000197635), score: 0.41 DPYSL3dihydropyrimidinase-like 3 (ENSG00000113657), score: -0.51 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (ENSG00000134109), score: 0.45 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.42 EGFRepidermal growth factor receptor (ENSG00000146648), score: 0.5 EHHADHenoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (ENSG00000113790), score: 0.42 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (ENSG00000158711), score: 0.51 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.46 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: 0.44 ERBB2IPerbb2 interacting protein (ENSG00000112851), score: 0.53 ERN1endoplasmic reticulum to nucleus signaling 1 (ENSG00000178607), score: 0.52 EVI5ecotropic viral integration site 5 (ENSG00000067208), score: 0.56 EXOC6exocyst complex component 6 (ENSG00000138190), score: 0.4 FABP2fatty acid binding protein 2, intestinal (ENSG00000145384), score: 0.87 FAM105Bfamily with sequence similarity 105, member B (ENSG00000154124), score: 0.48 FAM116Bfamily with sequence similarity 116, member B (ENSG00000205593), score: -0.5 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: -0.48 FAM160B1family with sequence similarity 160, member B1 (ENSG00000151553), score: 0.42 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: 0.51 FAM176Afamily with sequence similarity 176, member A (ENSG00000115363), score: 0.43 FAM59Afamily with sequence similarity 59, member A (ENSG00000141441), score: 0.43 FAM69Bfamily with sequence similarity 69, member B (ENSG00000165716), score: -0.5 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: 0.42 FBXL4F-box and leucine-rich repeat protein 4 (ENSG00000112234), score: 0.46 FBXW8F-box and WD repeat domain containing 8 (ENSG00000174989), score: 0.45 FCHO2FCH domain only 2 (ENSG00000157107), score: 0.48 FEZ1fasciculation and elongation protein zeta 1 (zygin I) (ENSG00000149557), score: -0.51 FRMD4BFERM domain containing 4B (ENSG00000114541), score: 0.4 FRRS1ferric-chelate reductase 1 (ENSG00000156869), score: 0.53 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (ENSG00000141429), score: 0.41 GALNTL1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1 (ENSG00000100626), score: -0.48 GAS2growth arrest-specific 2 (ENSG00000148935), score: 0.55 GBA2glucosidase, beta (bile acid) 2 (ENSG00000070610), score: -0.59 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.53 GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: 0.42 GGPS1geranylgeranyl diphosphate synthase 1 (ENSG00000152904), score: -0.54 GJB2gap junction protein, beta 2, 26kDa (ENSG00000165474), score: 0.67 GM2AGM2 ganglioside activator (ENSG00000196743), score: 0.6 GNA13guanine nucleotide binding protein (G protein), alpha 13 (ENSG00000120063), score: 0.42 GNEglucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase (ENSG00000159921), score: 0.53 GNMTglycine N-methyltransferase (ENSG00000124713), score: 0.44 GPLD1glycosylphosphatidylinositol specific phospholipase D1 (ENSG00000112293), score: 0.43 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.56 GPR97G protein-coupled receptor 97 (ENSG00000182885), score: 0.47 GRPEL2GrpE-like 2, mitochondrial (E. coli) (ENSG00000164284), score: 0.51 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: 0.46 H1FXH1 histone family, member X (ENSG00000184897), score: -0.87 H6PDhexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (ENSG00000049239), score: 0.62 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: 0.42 HDDC2HD domain containing 2 (ENSG00000111906), score: -0.51 HDGFRP3hepatoma-derived growth factor, related protein 3 (ENSG00000166503), score: -0.48 HELBhelicase (DNA) B (ENSG00000127311), score: 0.5 HERPUD1homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 (ENSG00000051108), score: 0.51 HEY1hairy/enhancer-of-split related with YRPW motif 1 (ENSG00000164683), score: -0.53 HGDhomogentisate 1,2-dioxygenase (ENSG00000113924), score: 0.45 HINFPhistone H4 transcription factor (ENSG00000172273), score: -0.48 HNF4Ahepatocyte nuclear factor 4, alpha (ENSG00000101076), score: 0.51 HNRNPRheterogeneous nuclear ribonucleoprotein R (ENSG00000125944), score: -0.6 IAH1isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) (ENSG00000134330), score: 0.51 IDH1isocitrate dehydrogenase 1 (NADP+), soluble (ENSG00000138413), score: 0.47 IFFO1intermediate filament family orphan 1 (ENSG00000010295), score: -0.59 IGHMBP2immunoglobulin mu binding protein 2 (ENSG00000132740), score: -0.48 INPP5Binositol polyphosphate-5-phosphatase, 75kDa (ENSG00000204084), score: 0.52 ITCHitchy E3 ubiquitin protein ligase homolog (mouse) (ENSG00000078747), score: 0.54 ITM2Aintegral membrane protein 2A (ENSG00000078596), score: -0.54 ITM2Bintegral membrane protein 2B (ENSG00000136156), score: 0.51 IYDiodotyrosine deiodinase (ENSG00000009765), score: 0.52 JAG2jagged 2 (ENSG00000184916), score: -0.47 KIAA0100KIAA0100 (ENSG00000007202), score: 0.43 KIAA2018KIAA2018 (ENSG00000176542), score: 0.55 KLF15Kruppel-like factor 15 (ENSG00000163884), score: 0.41 KLHDC2kelch domain containing 2 (ENSG00000165516), score: -0.57 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (ENSG00000117009), score: 0.45 KRT80keratin 80 (ENSG00000167767), score: 0.47 LAMP2lysosomal-associated membrane protein 2 (ENSG00000005893), score: 0.59 LARP4BLa ribonucleoprotein domain family, member 4B (ENSG00000107929), score: 0.51 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.5 LDHDlactate dehydrogenase D (ENSG00000166816), score: 0.47 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: 0.4 LIFRleukemia inhibitory factor receptor alpha (ENSG00000113594), score: 0.69 LMBRD2LMBR1 domain containing 2 (ENSG00000164187), score: 0.51 LPGAT1lysophosphatidylglycerol acyltransferase 1 (ENSG00000123684), score: 0.48 LRIT2leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 (ENSG00000204033), score: 0.47 LRRC20leucine rich repeat containing 20 (ENSG00000172731), score: -0.47 MAN1C1mannosidase, alpha, class 1C, member 1 (ENSG00000117643), score: -0.48 MAN2A1mannosidase, alpha, class 2A, member 1 (ENSG00000112893), score: 0.51 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.55 MAP3K5mitogen-activated protein kinase kinase kinase 5 (ENSG00000197442), score: 0.43 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.61 MARCH6membrane-associated ring finger (C3HC4) 6 (ENSG00000145495), score: 0.47 MBIPMAP3K12 binding inhibitory protein 1 (ENSG00000151332), score: -0.58 MCCC1methylcrotonoyl-CoA carboxylase 1 (alpha) (ENSG00000078070), score: 0.47 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.6 MCM3APminichromosome maintenance complex component 3 associated protein (ENSG00000160294), score: -0.51 MCRS1microspherule protein 1 (ENSG00000187778), score: -0.63 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.67 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.53 MEP1Ameprin A, alpha (PABA peptide hydrolase) (ENSG00000112818), score: 0.7 MEP1Bmeprin A, beta (ENSG00000141434), score: 0.72 MIA3melanoma inhibitory activity family, member 3 (ENSG00000154305), score: 0.56 MICAL1microtubule associated monoxygenase, calponin and LIM domain containing 1 (ENSG00000135596), score: -0.47 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.51 MIPOL1mirror-image polydactyly 1 (ENSG00000151338), score: 0.6 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.48 MOCOSmolybdenum cofactor sulfurase (ENSG00000075643), score: 0.46 MUTmethylmalonyl CoA mutase (ENSG00000146085), score: 0.48 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: -0.47 NADKNAD kinase (ENSG00000008130), score: 0.45 NAPEPLDN-acyl phosphatidylethanolamine phospholipase D (ENSG00000161048), score: -0.5 NDRG4NDRG family member 4 (ENSG00000103034), score: -0.47 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.62 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.62 NHLRC2NHL repeat containing 2 (ENSG00000196865), score: 0.44 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.51 NR1H4nuclear receptor subfamily 1, group H, member 4 (ENSG00000012504), score: 0.52 NSDHLNAD(P) dependent steroid dehydrogenase-like (ENSG00000147383), score: 0.44 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.68 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: 0.42 NUAK2NUAK family, SNF1-like kinase, 2 (ENSG00000163545), score: 0.45 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.66 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: 0.42 OSGIN2oxidative stress induced growth inhibitor family member 2 (ENSG00000164823), score: -0.55 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.45 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.64 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.47 PAHphenylalanine hydroxylase (ENSG00000171759), score: 0.53 PAK1IP1PAK1 interacting protein 1 (ENSG00000111845), score: 0.5 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: -0.49 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.53 PAPSS23'-phosphoadenosine 5'-phosphosulfate synthase 2 (ENSG00000198682), score: 0.55 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.42 PARD3par-3 partitioning defective 3 homolog (C. elegans) (ENSG00000148498), score: 0.46 PCBD2pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 (ENSG00000132570), score: 0.43 PCK1phosphoenolpyruvate carboxykinase 1 (soluble) (ENSG00000124253), score: 0.45 PCTPphosphatidylcholine transfer protein (ENSG00000141179), score: 0.42 PDP1pyruvate dehyrogenase phosphatase catalytic subunit 1 (ENSG00000164951), score: -0.47 PDS5APDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) (ENSG00000121892), score: 0.41 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.58 PEX16peroxisomal biogenesis factor 16 (ENSG00000121680), score: 0.42 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.45 PIK3IP1phosphoinositide-3-kinase interacting protein 1 (ENSG00000100100), score: -0.55 PLAAphospholipase A2-activating protein (ENSG00000137055), score: 0.43 PLK3polo-like kinase 3 (ENSG00000173846), score: 0.64 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.5 POLR1Cpolymerase (RNA) I polypeptide C, 30kDa (ENSG00000171453), score: 0.43 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (ENSG00000113356), score: 0.44 PPM1Bprotein phosphatase, Mg2+/Mn2+ dependent, 1B (ENSG00000138032), score: 0.41 PPP1R15Bprotein phosphatase 1, regulatory (inhibitor) subunit 15B (ENSG00000158615), score: 0.56 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (ENSG00000132825), score: -0.56 PPP1R7protein phosphatase 1, regulatory (inhibitor) subunit 7 (ENSG00000115685), score: -0.53 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha (ENSG00000066027), score: 0.53 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: -0.66 PRKAB2protein kinase, AMP-activated, beta 2 non-catalytic subunit (ENSG00000131791), score: -0.52 PRKAR1Bprotein kinase, cAMP-dependent, regulatory, type I, beta (ENSG00000188191), score: -0.48 PRKD3protein kinase D3 (ENSG00000115825), score: 0.65 PRLRprolactin receptor (ENSG00000113494), score: 0.71 PROSCproline synthetase co-transcribed homolog (bacterial) (ENSG00000147471), score: 0.42 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: -0.48 PSMA4proteasome (prosome, macropain) subunit, alpha type, 4 (ENSG00000041357), score: 0.53 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (ENSG00000197170), score: 0.44 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (ENSG00000152229), score: 0.43 PTERphosphotriesterase related (ENSG00000165983), score: 0.49 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: 0.44 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.48 RDXradixin (ENSG00000137710), score: 0.52 RFESDRieske (Fe-S) domain containing (ENSG00000175449), score: 0.44 RHOBTB3Rho-related BTB domain containing 3 (ENSG00000164292), score: -0.48 RNF139ring finger protein 139 (ENSG00000170881), score: 0.46 RNF34ring finger protein 34 (ENSG00000170633), score: -0.55 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.47 RXRAretinoid X receptor, alpha (ENSG00000186350), score: 0.41 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (ENSG00000211456), score: 0.44 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.52 SARDHsarcosine dehydrogenase (ENSG00000123453), score: 0.56 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: -0.5 SCCPDHsaccharopine dehydrogenase (putative) (ENSG00000143653), score: -0.73 SCLYselenocysteine lyase (ENSG00000132330), score: 0.48 SDR42E1short chain dehydrogenase/reductase family 42E, member 1 (ENSG00000184860), score: 0.5 SEC63SEC63 homolog (S. cerevisiae) (ENSG00000025796), score: 0.4 SEP1515 kDa selenoprotein (ENSG00000183291), score: 0.54 SEPSECSSep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (ENSG00000109618), score: 0.54 SEPT6septin 6 (ENSG00000125354), score: -0.58 SERINC3serine incorporator 3 (ENSG00000132824), score: 0.51 SFMBT2Scm-like with four mbt domains 2 (ENSG00000198879), score: -0.56 SFXN1sideroflexin 1 (ENSG00000164466), score: 0.42 SGCBsarcoglycan, beta (43kDa dystrophin-associated glycoprotein) (ENSG00000163069), score: -0.54 SGPP1sphingosine-1-phosphate phosphatase 1 (ENSG00000126821), score: 0.53 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: -0.58 SIRT3sirtuin 3 (ENSG00000142082), score: 0.46 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.72 SLC16A10solute carrier family 16, member 10 (aromatic amino acid transporter) (ENSG00000112394), score: 0.57 SLC16A12solute carrier family 16, member 12 (monocarboxylic acid transporter 12) (ENSG00000152779), score: 0.53 SLC16A7solute carrier family 16, member 7 (monocarboxylic acid transporter 2) (ENSG00000118596), score: 0.41 SLC17A8solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 (ENSG00000179520), score: 0.64 SLC18A1solute carrier family 18 (vesicular monoamine), member 1 (ENSG00000036565), score: 0.73 SLC25A10solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 (ENSG00000183048), score: 0.41 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: 0.58 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (ENSG00000100372), score: 0.55 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: -0.49 SLC2A2solute carrier family 2 (facilitated glucose transporter), member 2 (ENSG00000163581), score: 0.48 SLC2A9solute carrier family 2 (facilitated glucose transporter), member 9 (ENSG00000109667), score: 0.43 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.46 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (ENSG00000169359), score: 0.45 SLC35D1solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 (ENSG00000116704), score: 0.52 SLC39A6solute carrier family 39 (zinc transporter), member 6 (ENSG00000141424), score: -0.55 SLC39A8solute carrier family 39 (zinc transporter), member 8 (ENSG00000138821), score: 0.45 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.41 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (ENSG00000103064), score: -0.6 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.49 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.7 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (ENSG00000100796), score: 0.4 SNAPC3small nuclear RNA activating complex, polypeptide 3, 50kDa (ENSG00000164975), score: -0.48 SNX11sorting nexin 11 (ENSG00000002919), score: -0.51 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.55 SQLEsqualene epoxidase (ENSG00000104549), score: 0.44 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.61 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (ENSG00000136840), score: -0.57 STARD5StAR-related lipid transfer (START) domain containing 5 (ENSG00000172345), score: 0.57 SUN1Sad1 and UNC84 domain containing 1 (ENSG00000164828), score: -0.5 TBCELtubulin folding cofactor E-like (ENSG00000154114), score: 0.55 TCEA2transcription elongation factor A (SII), 2 (ENSG00000171703), score: -0.49 TCN2transcobalamin II (ENSG00000185339), score: 0.44 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: -0.5 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.49 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (ENSG00000100726), score: -0.52 TGDSTDP-glucose 4,6-dehydratase (ENSG00000088451), score: 0.54 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (ENSG00000135966), score: 0.53 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: -0.49 TLCD1TLC domain containing 1 (ENSG00000160606), score: 0.42 TM2D2TM2 domain containing 2 (ENSG00000169490), score: 0.51 TM9SF2transmembrane 9 superfamily member 2 (ENSG00000125304), score: 0.47 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: 0.53 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.55 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.55 TMEM14Atransmembrane protein 14A (ENSG00000096092), score: -0.48 TMEM195transmembrane protein 195 (ENSG00000187546), score: 0.46 TMEM20transmembrane protein 20 (ENSG00000176273), score: 0.59 TMEM64transmembrane protein 64 (ENSG00000180694), score: 0.57 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.41 TNIKTRAF2 and NCK interacting kinase (ENSG00000154310), score: -0.52 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: 0.42 TSPAN3tetraspanin 3 (ENSG00000140391), score: -0.65 TSR1TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) (ENSG00000167721), score: 0.48 TTC36tetratricopeptide repeat domain 36 (ENSG00000172425), score: 0.5 TTC39Ctetratricopeptide repeat domain 39C (ENSG00000168234), score: 0.53 TTPAtocopherol (alpha) transfer protein (ENSG00000137561), score: 0.42 TUBE1tubulin, epsilon 1 (ENSG00000074935), score: -0.51 TXNDC15thioredoxin domain containing 15 (ENSG00000113621), score: 0.42 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: 0.5 TXNRD1thioredoxin reductase 1 (ENSG00000198431), score: 0.45 UBLCP1ubiquitin-like domain containing CTD phosphatase 1 (ENSG00000164332), score: 0.42 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.56 UBTD2ubiquitin domain containing 2 (ENSG00000168246), score: -0.57 VASH1vasohibin 1 (ENSG00000071246), score: -0.55 VEZTvezatin, adherens junctions transmembrane protein (ENSG00000028203), score: -0.47 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: 0.49 WDR36WD repeat domain 36 (ENSG00000134987), score: 0.54 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.64 WRBtryptophan rich basic protein (ENSG00000182093), score: -0.57 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.61 ZBTB43zinc finger and BTB domain containing 43 (ENSG00000169155), score: 0.41 ZC3H12Czinc finger CCCH-type containing 12C (ENSG00000149289), score: 0.41 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.7 ZDHHC6zinc finger, DHHC-type containing 6 (ENSG00000023041), score: 0.48 ZMPSTE24zinc metallopeptidase (STE24 homolog, S. cerevisiae) (ENSG00000084073), score: 0.44 ZNF423zinc finger protein 423 (ENSG00000102935), score: -0.49 ZNF518Bzinc finger protein 518B (ENSG00000178163), score: -0.53 ZNF750zinc finger protein 750 (ENSG00000141579), score: 1
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |