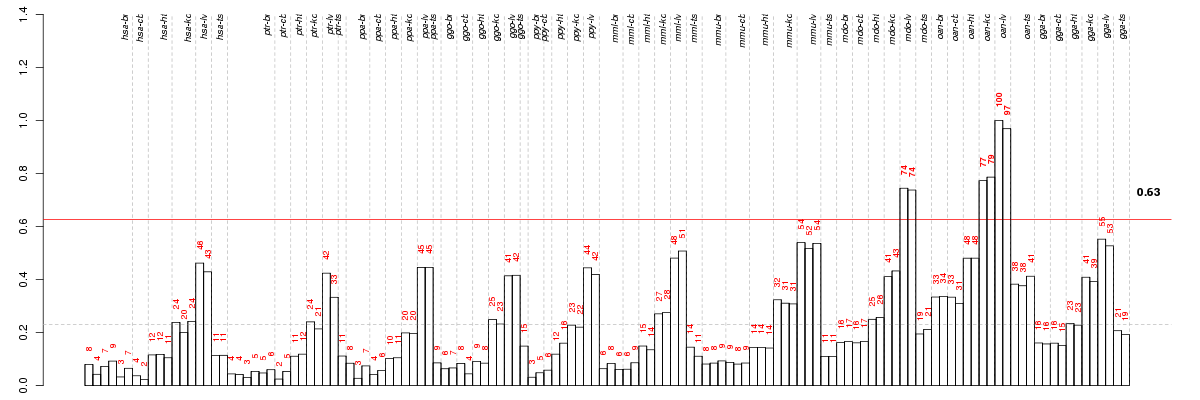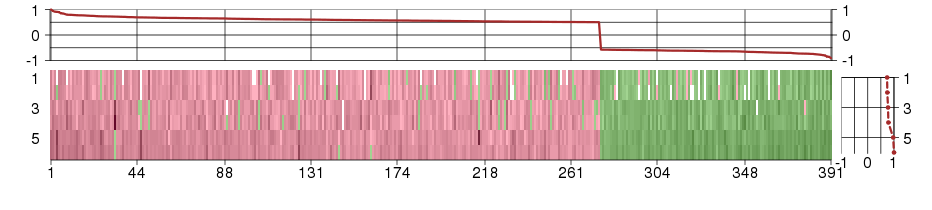

Under-expression is coded with green,
over-expression with red color.

sulfur amino acid metabolic process
The chemical reactions and pathways involving amino acids containing sulfur, comprising cysteine, homocysteine, methionine and selenocysteine.
sulfur metabolic process
The chemical reactions and pathways involving the nonmetallic element sulfur or compounds that contain sulfur, such as the amino acids methionine and cysteine or the tripeptide glutathione.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
lipid localization
Any process by which a lipid is transported to, or maintained in, a specific location.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
macromolecule localization
Any process by which a macromolecule is transported to, or maintained in, a specific location.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
sulfur amino acid metabolic process
The chemical reactions and pathways involving amino acids containing sulfur, comprising cysteine, homocysteine, methionine and selenocysteine.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
lytic vacuole
A vacuole that is maintained at an acidic pH and which contains degradative enzymes, including a wide variety of acid hydrolases.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
lysosome
A small lytic vacuole that has cell cycle-independent morphology and is found in most animal cells and that contains a variety of hydrolases, most of which have their maximal activities in the pH range 5-6. The contained enzymes display latency if properly isolated. About 40 different lysosomal hydrolases are known and lysosomes have a great variety of morphologies and functions.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
organelle membrane
The lipid bilayer surrounding an organelle.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00270 | 6.550e-03 | 0.9156 | 6 | 11 | Cysteine and methionine metabolism |
| 01100 | 1.988e-02 | 29.38 | 46 | 353 | Metabolic pathways |
AARS2alanyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000124608), score: 0.6 ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (ENSG00000165029), score: 0.64 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (ENSG00000154265), score: -0.69 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.56 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.91 ACOT12acyl-CoA thioesterase 12 (ENSG00000172497), score: 0.68 ACSL5acyl-CoA synthetase long-chain family member 5 (ENSG00000197142), score: 0.55 ACSS3acyl-CoA synthetase short-chain family member 3 (ENSG00000111058), score: 0.57 ACVR1Cactivin A receptor, type IC (ENSG00000123612), score: 0.51 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.71 ADAadenosine deaminase (ENSG00000196839), score: 0.53 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.63 ADPGKADP-dependent glucokinase (ENSG00000159322), score: 0.69 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.61 AK2adenylate kinase 2 (ENSG00000004455), score: 0.68 AKAP12A kinase (PRKA) anchor protein 12 (ENSG00000131016), score: -0.63 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.85 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.55 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.66 AMDHD1amidohydrolase domain containing 1 (ENSG00000139344), score: 0.74 AMOTL1angiomotin like 1 (ENSG00000166025), score: -0.63 ANAPC2anaphase promoting complex subunit 2 (ENSG00000176248), score: -0.69 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.64 ANKLE2ankyrin repeat and LEM domain containing 2 (ENSG00000176915), score: -0.62 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: -0.63 AP3M2adaptor-related protein complex 3, mu 2 subunit (ENSG00000070718), score: -0.64 ARCN1archain 1 (ENSG00000095139), score: 0.61 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: 0.61 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: 0.66 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.68 ARL5BADP-ribosylation factor-like 5B (ENSG00000165997), score: 0.65 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.53 ASAH1N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ENSG00000104763), score: 0.58 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ENSG00000153317), score: -0.59 ASH2Lash2 (absent, small, or homeotic)-like (Drosophila) (ENSG00000129691), score: -0.62 ATF6activating transcription factor 6 (ENSG00000118217), score: 0.52 ATG13ATG13 autophagy related 13 homolog (S. cerevisiae) (ENSG00000175224), score: 0.55 ATP8B1ATPase, aminophospholipid transporter, class I, type 8B, member 1 (ENSG00000081923), score: 0.52 ATRNattractin (ENSG00000088812), score: 0.51 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.85 AZI15-azacytidine induced 1 (ENSG00000141577), score: -0.64 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (ENSG00000158470), score: -0.7 BAT2L1HLA-B associated transcript 2-like 1 (ENSG00000130723), score: -0.6 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (ENSG00000083123), score: 0.61 BDKRB1bradykinin receptor B1 (ENSG00000100739), score: 0.77 BLMHbleomycin hydrolase (ENSG00000108578), score: 0.53 BMP2bone morphogenetic protein 2 (ENSG00000125845), score: 0.67 BTKBruton agammaglobulinemia tyrosine kinase (ENSG00000010671), score: 0.54 BTRCbeta-transducin repeat containing (ENSG00000166167), score: -0.63 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.58 C16orf7chromosome 16 open reading frame 7 (ENSG00000075399), score: -0.61 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: -0.68 C18orf8chromosome 18 open reading frame 8 (ENSG00000141452), score: 0.61 C1orf27chromosome 1 open reading frame 27 (ENSG00000157181), score: 0.57 C20orf4chromosome 20 open reading frame 4 (ENSG00000131043), score: 0.6 C4orf34chromosome 4 open reading frame 34 (ENSG00000163683), score: 0.59 C5complement component 5 (ENSG00000106804), score: 0.66 C5orf28chromosome 5 open reading frame 28 (ENSG00000151881), score: 0.51 C5orf43chromosome 5 open reading frame 43 (ENSG00000188725), score: 0.6 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: 0.6 C8orf40chromosome 8 open reading frame 40 (ENSG00000176209), score: -0.59 C9orf91chromosome 9 open reading frame 91 (ENSG00000157693), score: 0.64 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.61 CAND2cullin-associated and neddylation-dissociated 2 (putative) (ENSG00000144712), score: -0.58 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.58 CBY1chibby homolog 1 (Drosophila) (ENSG00000100211), score: -0.64 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: 0.66 CCDC127coiled-coil domain containing 127 (ENSG00000164366), score: -0.62 CCDC28Acoiled-coil domain containing 28A (ENSG00000024862), score: -0.61 CCNHcyclin H (ENSG00000134480), score: -0.65 CD82CD82 molecule (ENSG00000085117), score: 0.72 CD86CD86 molecule (ENSG00000114013), score: 0.52 CDH1cadherin 1, type 1, E-cadherin (epithelial) (ENSG00000039068), score: 0.51 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.64 CDO1cysteine dioxygenase, type I (ENSG00000129596), score: 0.65 CEPT1choline/ethanolamine phosphotransferase 1 (ENSG00000134255), score: 0.67 CHRNA9cholinergic receptor, nicotinic, alpha 9 (ENSG00000174343), score: 0.53 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.63 CIRBPcold inducible RNA binding protein (ENSG00000099622), score: -0.62 CLCN4chloride channel 4 (ENSG00000073464), score: -0.59 CLDN1claudin 1 (ENSG00000163347), score: 0.6 CLIP1CAP-GLY domain containing linker protein 1 (ENSG00000130779), score: -0.64 CLK4CDC-like kinase 4 (ENSG00000113240), score: -0.66 CLPXClpX caseinolytic peptidase X homolog (E. coli) (ENSG00000166855), score: 0.73 COG4component of oligomeric golgi complex 4 (ENSG00000103051), score: -0.64 COL4A3BPcollagen, type IV, alpha 3 (Goodpasture antigen) binding protein (ENSG00000113163), score: 0.63 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -0.61 COPB1coatomer protein complex, subunit beta 1 (ENSG00000129083), score: 0.58 CREB3L3cAMP responsive element binding protein 3-like 3 (ENSG00000060566), score: 0.51 CRLS1cardiolipin synthase 1 (ENSG00000088766), score: 0.63 CSE1LCSE1 chromosome segregation 1-like (yeast) (ENSG00000124207), score: -0.65 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.73 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.65 CUEDC2CUE domain containing 2 (ENSG00000107874), score: -0.85 CXXC5CXXC finger 5 (ENSG00000171604), score: -0.62 CYB5R2cytochrome b5 reductase 2 (ENSG00000166394), score: 0.63 CYP2R1cytochrome P450, family 2, subfamily R, polypeptide 1 (ENSG00000186104), score: 0.69 CYP39A1cytochrome P450, family 39, subfamily A, polypeptide 1 (ENSG00000146233), score: 0.64 CYSLTR1cysteinyl leukotriene receptor 1 (ENSG00000173198), score: 0.67 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.57 DDA1DET1 and DDB1 associated 1 (ENSG00000130311), score: -0.6 DENND1BDENN/MADD domain containing 1B (ENSG00000213047), score: 0.66 DENND5ADENN/MADD domain containing 5A (ENSG00000184014), score: -0.72 DERL1Der1-like domain family, member 1 (ENSG00000136986), score: 0.6 DERL3Der1-like domain family, member 3 (ENSG00000099958), score: 0.51 DHDPSLdihydrodipicolinate synthase-like, mitochondrial (ENSG00000241935), score: 0.58 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.77 DIRC2disrupted in renal carcinoma 2 (ENSG00000138463), score: 0.73 DNAJC22DnaJ (Hsp40) homolog, subfamily C, member 22 (ENSG00000178401), score: 0.56 DOCK9dedicator of cytokinesis 9 (ENSG00000088387), score: -0.59 DPP8dipeptidyl-peptidase 8 (ENSG00000074603), score: 0.6 DPYSdihydropyrimidinase (ENSG00000147647), score: 0.51 DRAM2DNA-damage regulated autophagy modulator 2 (ENSG00000156171), score: 0.52 E2F5E2F transcription factor 5, p130-binding (ENSG00000133740), score: 0.51 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.79 EEF1E1eukaryotic translation elongation factor 1 epsilon 1 (ENSG00000124802), score: -0.62 EGFRepidermal growth factor receptor (ENSG00000146648), score: 0.52 EPB41L2erythrocyte membrane protein band 4.1-like 2 (ENSG00000079819), score: -0.6 EPHA1EPH receptor A1 (ENSG00000146904), score: 0.56 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (ENSG00000143819), score: 0.53 EPN2epsin 2 (ENSG00000072134), score: -0.66 EREGepiregulin (ENSG00000124882), score: 0.53 ERLIN1ER lipid raft associated 1 (ENSG00000107566), score: 0.53 ERP44endoplasmic reticulum protein 44 (ENSG00000023318), score: 0.51 ESR1estrogen receptor 1 (ENSG00000091831), score: 0.62 ETNK2ethanolamine kinase 2 (ENSG00000143845), score: 0.62 EXOC8exocyst complex component 8 (ENSG00000116903), score: 0.61 F2coagulation factor II (thrombin) (ENSG00000180210), score: 0.63 F9coagulation factor IX (ENSG00000101981), score: 0.52 FAF2Fas associated factor family member 2 (ENSG00000113194), score: 0.52 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.59 FAM171A1family with sequence similarity 171, member A1 (ENSG00000148468), score: -0.64 FAM3Cfamily with sequence similarity 3, member C (ENSG00000196937), score: -0.65 FAM3Dfamily with sequence similarity 3, member D (ENSG00000198643), score: 0.59 FAM63Bfamily with sequence similarity 63, member B (ENSG00000128923), score: 0.64 FAM69Bfamily with sequence similarity 69, member B (ENSG00000165716), score: -0.6 FAM82Bfamily with sequence similarity 82, member B (ENSG00000176623), score: 0.52 FBXO4F-box protein 4 (ENSG00000151876), score: 0.66 FETUBfetuin B (ENSG00000090512), score: 0.64 FHfumarate hydratase (ENSG00000091483), score: 0.59 FSHBfollicle stimulating hormone, beta polypeptide (ENSG00000131808), score: 0.72 FXR1fragile X mental retardation, autosomal homolog 1 (ENSG00000114416), score: -0.6 FZD2frizzled homolog 2 (Drosophila) (ENSG00000180340), score: 0.63 GALK2galactokinase 2 (ENSG00000156958), score: 0.52 GAR1GAR1 ribonucleoprotein homolog (yeast) (ENSG00000109534), score: 0.66 GAS6growth arrest-specific 6 (ENSG00000183087), score: -0.79 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.77 GGHgamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (ENSG00000137563), score: 0.61 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: 0.51 GLO1glyoxalase I (ENSG00000124767), score: 0.54 GNB5guanine nucleotide binding protein (G protein), beta 5 (ENSG00000069966), score: -0.57 GNMTglycine N-methyltransferase (ENSG00000124713), score: 0.67 GOLIM4golgi integral membrane protein 4 (ENSG00000173905), score: 0.59 GOT1glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (ENSG00000120053), score: 0.62 GPHNgephyrin (ENSG00000171723), score: 0.61 GPN1GPN-loop GTPase 1 (ENSG00000198522), score: 0.62 GTF2E1general transcription factor IIE, polypeptide 1, alpha 56kDa (ENSG00000153767), score: 0.65 HALhistidine ammonia-lyase (ENSG00000084110), score: 0.62 HEBP1heme binding protein 1 (ENSG00000013583), score: 0.61 HELQhelicase, POLQ-like (ENSG00000163312), score: 0.51 HGDhomogentisate 1,2-dioxygenase (ENSG00000113924), score: 0.56 HM13histocompatibility (minor) 13 (ENSG00000101294), score: 0.72 HMGXB4HMG box domain containing 4 (ENSG00000100281), score: 0.59 HNF4Ghepatocyte nuclear factor 4, gamma (ENSG00000164749), score: 0.62 HTThuntingtin (ENSG00000197386), score: -0.68 HYIhydroxypyruvate isomerase (putative) (ENSG00000178922), score: 0.62 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (ENSG00000005700), score: 0.66 IDEinsulin-degrading enzyme (ENSG00000119912), score: 0.58 IGF1Rinsulin-like growth factor 1 receptor (ENSG00000140443), score: -0.58 IGFBP1insulin-like growth factor binding protein 1 (ENSG00000146678), score: 0.66 ITCHitchy E3 ubiquitin protein ligase homolog (mouse) (ENSG00000078747), score: 0.56 ITGA4integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) (ENSG00000115232), score: 0.73 ITGB1BP1integrin beta 1 binding protein 1 (ENSG00000119185), score: -0.59 IYDiodotyrosine deiodinase (ENSG00000009765), score: 0.79 KCTD6potassium channel tetramerisation domain containing 6 (ENSG00000168301), score: 0.52 KDM1Blysine (K)-specific demethylase 1B (ENSG00000165097), score: 0.56 KIAA0090KIAA0090 (ENSG00000127463), score: 0.67 KIAA0226KIAA0226 (ENSG00000145016), score: -0.59 KIAA0247KIAA0247 (ENSG00000100647), score: 0.58 KIAA0664KIAA0664 (ENSG00000132361), score: 0.58 KIAA0907KIAA0907 (ENSG00000132680), score: 0.59 KIAA1432KIAA1432 (ENSG00000107036), score: 0.5 KLHDC8Bkelch domain containing 8B (ENSG00000185909), score: -0.73 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.91 KLHL8kelch-like 8 (Drosophila) (ENSG00000145332), score: 0.65 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (ENSG00000117009), score: 0.69 LAMB3laminin, beta 3 (ENSG00000196878), score: 0.59 LEAP2liver expressed antimicrobial peptide 2 (ENSG00000164406), score: 0.7 LECT2leukocyte cell-derived chemotaxin 2 (ENSG00000145826), score: 0.69 LIPClipase, hepatic (ENSG00000166035), score: 0.67 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: -0.67 LOC100292021similar to thioredoxin peroxidase (ENSG00000123131), score: 0.67 LPCAT3lysophosphatidylcholine acyltransferase 3 (ENSG00000111684), score: 0.57 LRATlecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) (ENSG00000121207), score: 0.52 LRIG3leucine-rich repeats and immunoglobulin-like domains 3 (ENSG00000139263), score: 0.75 LRP11low density lipoprotein receptor-related protein 11 (ENSG00000120256), score: -0.7 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.69 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.73 LRRC58leucine rich repeat containing 58 (ENSG00000163428), score: 0.72 LY75lymphocyte antigen 75 (ENSG00000054219), score: 0.67 M6PRmannose-6-phosphate receptor (cation dependent) (ENSG00000003056), score: 0.53 MAGT1magnesium transporter 1 (ENSG00000102158), score: 0.56 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: 0.57 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: 0.65 MAP2K5mitogen-activated protein kinase kinase 5 (ENSG00000137764), score: -0.74 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (ENSG00000138834), score: -0.66 MARK3MAP/microtubule affinity-regulating kinase 3 (ENSG00000075413), score: -0.61 MARVELD3MARVEL domain containing 3 (ENSG00000140832), score: 0.58 MAT1Amethionine adenosyltransferase I, alpha (ENSG00000151224), score: 0.55 MBD3methyl-CpG binding domain protein 3 (ENSG00000071655), score: -0.62 MBNL2muscleblind-like 2 (Drosophila) (ENSG00000139793), score: -0.85 MED23mediator complex subunit 23 (ENSG00000112282), score: 0.62 MKLN1muskelin 1, intracellular mediator containing kelch motifs (ENSG00000128585), score: 0.65 MLLT6myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 (ENSG00000108292), score: -0.61 MLXMAX-like protein X (ENSG00000108788), score: 0.52 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.54 MOCS1molybdenum cofactor synthesis 1 (ENSG00000124615), score: 0.52 MPDZmultiple PDZ domain protein (ENSG00000107186), score: -0.7 MPZL2myelin protein zero-like 2 (ENSG00000149573), score: 0.57 MRPL12mitochondrial ribosomal protein L12 (ENSG00000183093), score: -0.61 MRPS30mitochondrial ribosomal protein S30 (ENSG00000112996), score: 0.6 MSRAmethionine sulfoxide reductase A (ENSG00000175806), score: 0.57 MTA1metastasis associated 1 (ENSG00000182979), score: -0.69 MTAPmethylthioadenosine phosphorylase (ENSG00000099810), score: 0.75 MTFMTmitochondrial methionyl-tRNA formyltransferase (ENSG00000103707), score: 0.53 MTMR2myotubularin related protein 2 (ENSG00000087053), score: -0.62 MTPAPmitochondrial poly(A) polymerase (ENSG00000107951), score: 0.57 MTTPmicrosomal triglyceride transfer protein (ENSG00000138823), score: 0.6 MUDENGMU-2/AP1M2 domain containing, death-inducing (ENSG00000053770), score: 0.55 MYNNmyoneurin (ENSG00000085274), score: 0.63 N6AMT2N-6 adenine-specific DNA methyltransferase 2 (putative) (ENSG00000150456), score: 0.51 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.76 NCSTNnicastrin (ENSG00000162736), score: 0.73 NELFnasal embryonic LHRH factor (ENSG00000165802), score: -0.62 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: 0.77 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.77 NRBP1nuclear receptor binding protein 1 (ENSG00000115216), score: -0.6 NT5DC25'-nucleotidase domain containing 2 (ENSG00000168268), score: -0.59 NT5E5'-nucleotidase, ecto (CD73) (ENSG00000135318), score: 0.79 OIT3oncoprotein induced transcript 3 (ENSG00000138315), score: 0.52 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: -0.61 OSTM1osteopetrosis associated transmembrane protein 1 (ENSG00000081087), score: 0.57 OTCornithine carbamoyltransferase (ENSG00000036473), score: 0.9 OXSR1oxidative-stress responsive 1 (ENSG00000172939), score: 0.56 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (ENSG00000122884), score: 0.57 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.75 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: -0.7 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.69 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: 0.52 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.6 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.54 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (ENSG00000102981), score: -0.61 PARP6poly (ADP-ribose) polymerase family, member 6 (ENSG00000137817), score: -0.78 PCK1phosphoenolpyruvate carboxykinase 1 (soluble) (ENSG00000124253), score: 0.5 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.7 PDE3Bphosphodiesterase 3B, cGMP-inhibited (ENSG00000152270), score: 0.53 PEMTphosphatidylethanolamine N-methyltransferase (ENSG00000133027), score: 0.56 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.78 PGM3phosphoglucomutase 3 (ENSG00000013375), score: 0.79 PHOSPHO1phosphatase, orphan 1 (ENSG00000173868), score: 0.54 PIAS1protein inhibitor of activated STAT, 1 (ENSG00000033800), score: -0.67 PIGXphosphatidylinositol glycan anchor biosynthesis, class X (ENSG00000163964), score: 0.6 PIP5K1Bphosphatidylinositol-4-phosphate 5-kinase, type I, beta (ENSG00000107242), score: -0.58 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (ENSG00000186111), score: -0.7 PITPNAphosphatidylinositol transfer protein, alpha (ENSG00000174238), score: -0.59 PLA2G12Bphospholipase A2, group XIIB (ENSG00000138308), score: 0.55 PLEKHF2pleckstrin homology domain containing, family F (with FYVE domain) member 2 (ENSG00000175895), score: 0.59 PM20D2peptidase M20 domain containing 2 (ENSG00000146281), score: 0.55 POFUT1protein O-fucosyltransferase 1 (ENSG00000101346), score: 0.58 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (ENSG00000132664), score: 0.52 PPEF2protein phosphatase, EF-hand calcium binding domain 2 (ENSG00000156194), score: 0.58 PPIL3peptidylprolyl isomerase (cyclophilin)-like 3 (ENSG00000240344), score: 0.56 PRKCDprotein kinase C, delta (ENSG00000163932), score: -0.59 PSEN2presenilin 2 (Alzheimer disease 4) (ENSG00000143801), score: 0.66 QPCTglutaminyl-peptide cyclotransferase (ENSG00000115828), score: 0.52 QSOX1quiescin Q6 sulfhydryl oxidase 1 (ENSG00000116260), score: -0.64 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.55 RAB33BRAB33B, member RAS oncogene family (ENSG00000172007), score: 0.67 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.54 RAD17RAD17 homolog (S. pombe) (ENSG00000152942), score: 0.51 RANBP3RAN binding protein 3 (ENSG00000031823), score: -0.74 RASA3RAS p21 protein activator 3 (ENSG00000185989), score: -0.6 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.61 RECKreversion-inducing-cysteine-rich protein with kazal motifs (ENSG00000122707), score: 0.57 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.53 RGS2regulator of G-protein signaling 2, 24kDa (ENSG00000116741), score: -0.73 RNF103ring finger protein 103 (ENSG00000239305), score: 0.5 RNF144Aring finger protein 144A (ENSG00000151692), score: -0.67 RPAINRPA interacting protein (ENSG00000129197), score: 0.54 SCAMP2secretory carrier membrane protein 2 (ENSG00000140497), score: 0.53 SDCCAG8serologically defined colon cancer antigen 8 (ENSG00000054282), score: -0.62 SERINC5serine incorporator 5 (ENSG00000164300), score: 0.66 SERPINC1serpin peptidase inhibitor, clade C (antithrombin), member 1 (ENSG00000117601), score: 0.6 SETD5SET domain containing 5 (ENSG00000168137), score: -0.69 SGSM2small G protein signaling modulator 2 (ENSG00000141258), score: -0.67 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: -0.64 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (ENSG00000204351), score: 0.51 SLC16A12solute carrier family 16, member 12 (monocarboxylic acid transporter 12) (ENSG00000152779), score: 0.55 SLC16A2solute carrier family 16, member 2 (monocarboxylic acid transporter 8) (ENSG00000147100), score: 0.61 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (ENSG00000119899), score: 0.7 SLC25A40solute carrier family 25, member 40 (ENSG00000075303), score: 0.51 SLC25A47solute carrier family 25, member 47 (ENSG00000140107), score: 0.57 SLC2A2solute carrier family 2 (facilitated glucose transporter), member 2 (ENSG00000163581), score: 0.52 SLC2A9solute carrier family 2 (facilitated glucose transporter), member 9 (ENSG00000109667), score: 0.71 SLC30A1solute carrier family 30 (zinc transporter), member 1 (ENSG00000170385), score: 0.67 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.64 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.73 SLC31A1solute carrier family 31 (copper transporters), member 1 (ENSG00000136868), score: 0.64 SLC38A4solute carrier family 38, member 4 (ENSG00000139209), score: 0.53 SLC39A9solute carrier family 39 (zinc transporter), member 9 (ENSG00000029364), score: 0.94 SLC40A1solute carrier family 40 (iron-regulated transporter), member 1 (ENSG00000138449), score: 0.59 SLC45A3solute carrier family 45, member 3 (ENSG00000158715), score: 0.56 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.76 SLC7A2solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 (ENSG00000003989), score: 0.51 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (ENSG00000103064), score: -0.67 SMYD4SET and MYND domain containing 4 (ENSG00000186532), score: -0.69 SNED1sushi, nidogen and EGF-like domains 1 (ENSG00000162804), score: -0.73 SNRNP48small nuclear ribonucleoprotein 48kDa (U11/U12) (ENSG00000168566), score: 0.57 SNX24sorting nexin 24 (ENSG00000064652), score: 0.65 SPATA5L1spermatogenesis associated 5-like 1 (ENSG00000171763), score: 0.57 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.5 SPTLC2serine palmitoyltransferase, long chain base subunit 2 (ENSG00000100596), score: 0.54 SRSF4serine/arginine-rich splicing factor 4 (ENSG00000116350), score: -0.59 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: -0.66 SS18synovial sarcoma translocation, chromosome 18 (ENSG00000141380), score: 0.66 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: 0.63 ST8SIA4ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (ENSG00000113532), score: 0.52 STARD5StAR-related lipid transfer (START) domain containing 5 (ENSG00000172345), score: 0.56 STK39serine threonine kinase 39 (ENSG00000198648), score: -0.61 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.62 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: 0.59 TARSL2threonyl-tRNA synthetase-like 2 (ENSG00000185418), score: -0.7 TATtyrosine aminotransferase (ENSG00000198650), score: 0.61 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (ENSG00000133138), score: 0.5 TCEA2transcription elongation factor A (SII), 2 (ENSG00000171703), score: -0.58 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.58 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.51 TLN2talin 2 (ENSG00000171914), score: -0.62 TMCO3transmembrane and coiled-coil domains 3 (ENSG00000150403), score: -0.74 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.7 TMEM33transmembrane protein 33 (ENSG00000109133), score: 0.68 TMEM38Atransmembrane protein 38A (ENSG00000072954), score: -0.71 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.77 TOM1L2target of myb1-like 2 (chicken) (ENSG00000175662), score: -0.64 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.73 TP53INP1tumor protein p53 inducible nuclear protein 1 (ENSG00000164938), score: 0.57 TRAM2translocation associated membrane protein 2 (ENSG00000065308), score: 0.58 TRIP13thyroid hormone receptor interactor 13 (ENSG00000071539), score: 0.56 TRNAU1APtRNA selenocysteine 1 associated protein 1 (ENSG00000180098), score: 0.51 TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: 0.57 TSKUtsukushi small leucine rich proteoglycan homolog (Xenopus laevis) (ENSG00000182704), score: 0.54 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: -0.62 TTC1tetratricopeptide repeat domain 1 (ENSG00000113312), score: -0.68 TTC36tetratricopeptide repeat domain 36 (ENSG00000172425), score: 0.55 TTPAtocopherol (alpha) transfer protein (ENSG00000137561), score: 0.59 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.64 TWISTNBTWIST neighbor (ENSG00000105849), score: 0.82 TWSG1twisted gastrulation homolog 1 (Drosophila) (ENSG00000128791), score: -0.73 TXNDC15thioredoxin domain containing 15 (ENSG00000113621), score: 0.62 UBA6ubiquitin-like modifier activating enzyme 6 (ENSG00000033178), score: 0.61 UBL3ubiquitin-like 3 (ENSG00000122042), score: 0.54 UBXN4UBX domain protein 4 (ENSG00000144224), score: 0.57 UHRF1BP1LUHRF1 binding protein 1-like (ENSG00000111647), score: 0.65 UIMC1ubiquitin interaction motif containing 1 (ENSG00000087206), score: -0.58 UPB1ureidopropionase, beta (ENSG00000100024), score: 0.55 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: 0.67 USP20ubiquitin specific peptidase 20 (ENSG00000136878), score: -0.8 UTP15UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) (ENSG00000164338), score: 0.68 VAT1vesicle amine transport protein 1 homolog (T. californica) (ENSG00000108828), score: 0.51 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.6 VLDLRvery low density lipoprotein receptor (ENSG00000147852), score: -0.58 VPS4Bvacuolar protein sorting 4 homolog B (S. cerevisiae) (ENSG00000119541), score: 0.66 WDFY2WD repeat and FYVE domain containing 2 (ENSG00000139668), score: 0.62 WDR3WD repeat domain 3 (ENSG00000065183), score: 0.56 WDR43WD repeat domain 43 (ENSG00000163811), score: 0.51 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.58 WDR6WD repeat domain 6 (ENSG00000178252), score: -0.64 WDR8WD repeat domain 8 (ENSG00000116213), score: -0.58 WNT9Awingless-type MMTV integration site family, member 9A (ENSG00000143816), score: 0.71 XKR9XK, Kell blood group complex subunit-related family, member 9 (ENSG00000221947), score: 1 XRCC6BP1XRCC6 binding protein 1 (ENSG00000166896), score: 0.65 YIPF5Yip1 domain family, member 5 (ENSG00000145817), score: 0.52 YTHDC2YTH domain containing 2 (ENSG00000047188), score: -0.76 ZBTB2zinc finger and BTB domain containing 2 (ENSG00000181472), score: 0.56 ZBTB49zinc finger and BTB domain containing 49 (ENSG00000168826), score: -0.6 ZC3H12Dzinc finger CCCH-type containing 12D (ENSG00000178199), score: 0.91 ZDHHC21zinc finger, DHHC-type containing 21 (ENSG00000175893), score: -0.58 ZMPSTE24zinc metallopeptidase (STE24 homolog, S. cerevisiae) (ENSG00000084073), score: 0.69 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.75 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.72 ZW10ZW10, kinetochore associated, homolog (Drosophila) (ENSG00000086827), score: 0.51
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| oan_kd_m_ca1 | oan | kd | m | _ |
| oan_kd_f_ca1 | oan | kd | f | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |