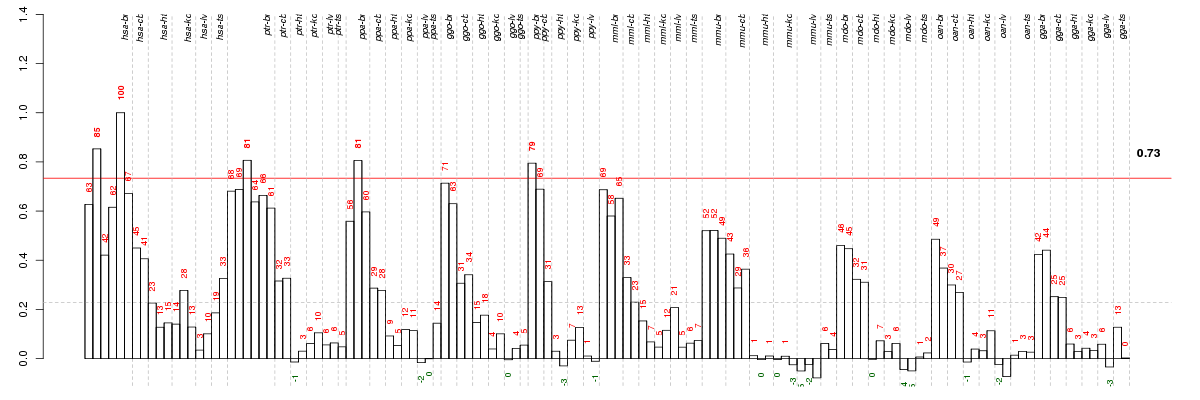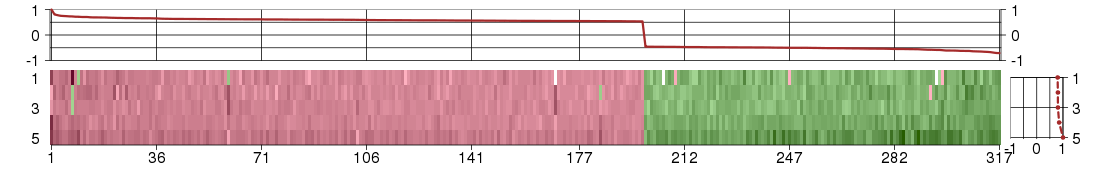

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
RNA catabolic process
The chemical reactions and pathways resulting in the breakdown of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
RNA catabolic process
The chemical reactions and pathways resulting in the breakdown of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.


ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: -0.62 ABRactive BCR-related gene (ENSG00000159842), score: 0.57 ACP6acid phosphatase 6, lysophosphatidic (ENSG00000162836), score: -0.48 ACTL6Aactin-like 6A (ENSG00000136518), score: -0.48 ADAMTS8ADAM metallopeptidase with thrombospondin type 1 motif, 8 (ENSG00000134917), score: 0.66 ADCY2adenylate cyclase 2 (brain) (ENSG00000078295), score: 0.55 AIMP1aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 (ENSG00000164022), score: -0.54 AKAP9A kinase (PRKA) anchor protein (yotiao) 9 (ENSG00000127914), score: -0.59 ALKBH4alkB, alkylation repair homolog 4 (E. coli) (ENSG00000160993), score: 0.54 ANKRD24ankyrin repeat domain 24 (ENSG00000089847), score: 0.6 AP3B1adaptor-related protein complex 3, beta 1 subunit (ENSG00000132842), score: -0.48 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: -0.53 ARCactivity-regulated cytoskeleton-associated protein (ENSG00000198576), score: 0.66 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: -0.61 ARFRP1ADP-ribosylation factor related protein 1 (ENSG00000101246), score: 0.55 ARHGAP12Rho GTPase activating protein 12 (ENSG00000165322), score: -0.51 ASB6ankyrin repeat and SOCS box-containing 6 (ENSG00000148331), score: 0.69 ATG4BATG4 autophagy related 4 homolog B (S. cerevisiae) (ENSG00000168397), score: 0.56 ATL2atlastin GTPase 2 (ENSG00000119787), score: -0.48 ATP13A2ATPase type 13A2 (ENSG00000159363), score: 0.63 ATP6V0BATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (ENSG00000117410), score: 0.55 BACH1BTB and CNC homology 1, basic leucine zipper transcription factor 1 (ENSG00000156273), score: -0.47 BAI1brain-specific angiogenesis inhibitor 1 (ENSG00000181790), score: 0.6 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (ENSG00000163930), score: 0.74 BCAS3breast carcinoma amplified sequence 3 (ENSG00000141376), score: 0.56 BEGAINbrain-enriched guanylate kinase-associated homolog (rat) (ENSG00000183092), score: 0.58 BTBD2BTB (POZ) domain containing 2 (ENSG00000133243), score: 0.61 C13orf36chromosome 13 open reading frame 36 (ENSG00000180440), score: 0.6 C1orf123chromosome 1 open reading frame 123 (ENSG00000162384), score: -0.46 C1orf128chromosome 1 open reading frame 128 (ENSG00000057757), score: 0.59 C1orf27chromosome 1 open reading frame 27 (ENSG00000157181), score: -0.47 C1orf55chromosome 1 open reading frame 55 (ENSG00000143751), score: -0.57 C20orf103chromosome 20 open reading frame 103 (ENSG00000125869), score: 0.54 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.62 C5orf41chromosome 5 open reading frame 41 (ENSG00000164463), score: -0.49 C8orf46chromosome 8 open reading frame 46 (ENSG00000169085), score: 0.6 C9orf16chromosome 9 open reading frame 16 (ENSG00000171159), score: 0.69 CA7carbonic anhydrase VII (ENSG00000168748), score: 0.61 CACNG3calcium channel, voltage-dependent, gamma subunit 3 (ENSG00000006116), score: 0.53 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 1 CAMK1Gcalcium/calmodulin-dependent protein kinase IG (ENSG00000008118), score: 0.62 CAMK2Acalcium/calmodulin-dependent protein kinase II alpha (ENSG00000070808), score: 0.6 CAPN9calpain 9 (ENSG00000135773), score: 0.67 CBLN4cerebellin 4 precursor (ENSG00000054803), score: 0.65 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: -0.55 CCDC3coiled-coil domain containing 3 (ENSG00000151468), score: 0.54 CCDC50coiled-coil domain containing 50 (ENSG00000152492), score: -0.46 CCM2cerebral cavernous malformation 2 (ENSG00000136280), score: 0.63 CD2APCD2-associated protein (ENSG00000198087), score: -0.49 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: 0.6 CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.61 CELF5CUGBP, Elav-like family member 5 (ENSG00000161082), score: 0.58 CETN3centrin, EF-hand protein, 3 (ENSG00000153140), score: -0.65 CHRM3cholinergic receptor, muscarinic 3 (ENSG00000133019), score: 0.6 CHRNA4cholinergic receptor, nicotinic, alpha 4 (ENSG00000101204), score: 0.58 CHST1carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 (ENSG00000175264), score: 0.6 CLCN7chloride channel 7 (ENSG00000103249), score: 0.57 CNIH3cornichon homolog 3 (Drosophila) (ENSG00000143786), score: 0.55 COBRA1cofactor of BRCA1 (ENSG00000188986), score: 0.55 COPB1coatomer protein complex, subunit beta 1 (ENSG00000129083), score: -0.53 CPNE4copine IV (ENSG00000196353), score: 0.54 CRTAC1cartilage acidic protein 1 (ENSG00000095713), score: 0.54 CTBP1C-terminal binding protein 1 (ENSG00000159692), score: 0.55 CTNND2catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein) (ENSG00000169862), score: 0.54 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: -0.46 DDX41DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (ENSG00000183258), score: 0.58 DGCR2DiGeorge syndrome critical region gene 2 (ENSG00000070413), score: 0.64 DGKZdiacylglycerol kinase, zeta 104kDa (ENSG00000149091), score: 0.69 DLGAP2discs, large (Drosophila) homolog-associated protein 2 (ENSG00000198010), score: 0.54 DLX1distal-less homeobox 1 (ENSG00000144355), score: 0.62 DPYSL4dihydropyrimidinase-like 4 (ENSG00000151640), score: 0.55 DSCAMDown syndrome cell adhesion molecule (ENSG00000171587), score: 0.54 EDC4enhancer of mRNA decapping 4 (ENSG00000038358), score: 0.56 EEA1early endosome antigen 1 (ENSG00000102189), score: -0.48 EIF3Eeukaryotic translation initiation factor 3, subunit E (ENSG00000104408), score: -0.48 EIF3Meukaryotic translation initiation factor 3, subunit M (ENSG00000149100), score: -0.47 EIF5Beukaryotic translation initiation factor 5B (ENSG00000158417), score: -0.5 ELFN2extracellular leucine-rich repeat and fibronectin type III domain containing 2 (ENSG00000166897), score: 0.62 ELMOD2ELMO/CED-12 domain containing 2 (ENSG00000179387), score: -0.71 ENC1ectodermal-neural cortex 1 (with BTB-like domain) (ENSG00000171617), score: 0.65 ENTPD6ectonucleoside triphosphate diphosphohydrolase 6 (putative) (ENSG00000197586), score: 0.62 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: -0.47 EPHB6EPH receptor B6 (ENSG00000106123), score: 0.68 ERI1exoribonuclease 1 (ENSG00000104626), score: -0.48 ETAA1Ewing tumor-associated antigen 1 (ENSG00000143971), score: -0.63 EXOC7exocyst complex component 7 (ENSG00000182473), score: 0.71 FAM102Afamily with sequence similarity 102, member A (ENSG00000167106), score: 0.6 FAM168Bfamily with sequence similarity 168, member B (ENSG00000152102), score: 0.59 FAM19A1family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 (ENSG00000183662), score: 0.67 FAM40Afamily with sequence similarity 40, member A (ENSG00000143093), score: 0.54 FAM5Bfamily with sequence similarity 5, member B (ENSG00000198797), score: 0.61 FAM60Afamily with sequence similarity 60, member A (ENSG00000139146), score: -0.48 FBLN7fibulin 7 (ENSG00000144152), score: 0.66 FBXL15F-box and leucine-rich repeat protein 15 (ENSG00000107872), score: 0.61 FBXL16F-box and leucine-rich repeat protein 16 (ENSG00000127585), score: 0.6 FBXL5F-box and leucine-rich repeat protein 5 (ENSG00000118564), score: -0.48 FBXO2F-box protein 2 (ENSG00000116661), score: 0.54 FBXO8F-box protein 8 (ENSG00000164117), score: -0.53 FCHO2FCH domain only 2 (ENSG00000157107), score: -0.54 FEZF2FEZ family zinc finger 2 (ENSG00000153266), score: 0.75 FGF22fibroblast growth factor 22 (ENSG00000070388), score: 0.6 FOXG1forkhead box G1 (ENSG00000176165), score: 0.68 GABPAGA binding protein transcription factor, alpha subunit 60kDa (ENSG00000154727), score: -0.63 GCFC1GC-rich sequence DNA-binding factor 1 (ENSG00000159086), score: -0.56 GJD4gap junction protein, delta 4, 40.1kDa (ENSG00000177291), score: 0.72 GNAZguanine nucleotide binding protein (G protein), alpha z polypeptide (ENSG00000128266), score: 0.55 GNB1guanine nucleotide binding protein (G protein), beta polypeptide 1 (ENSG00000078369), score: 0.61 GNL2guanine nucleotide binding protein-like 2 (nucleolar) (ENSG00000134697), score: -0.49 GOLGA4golgin A4 (ENSG00000144674), score: -0.69 GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.78 GPR176G protein-coupled receptor 176 (ENSG00000166073), score: 0.61 GRID1glutamate receptor, ionotropic, delta 1 (ENSG00000182771), score: 0.57 GRIN1glutamate receptor, ionotropic, N-methyl D-aspartate 1 (ENSG00000176884), score: 0.55 GRK6G protein-coupled receptor kinase 6 (ENSG00000198055), score: 0.56 GSG1LGSG1-like (ENSG00000169181), score: 0.56 GTF2A2general transcription factor IIA, 2, 12kDa (ENSG00000140307), score: -0.52 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: -0.49 HAT1histone acetyltransferase 1 (ENSG00000128708), score: -0.64 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: -0.48 HGShepatocyte growth factor-regulated tyrosine kinase substrate (ENSG00000185359), score: 0.62 HIBCH3-hydroxyisobutyryl-CoA hydrolase (ENSG00000198130), score: -0.49 HMBOX1homeobox containing 1 (ENSG00000147421), score: -0.49 HMGXB3HMG box domain containing 3 (ENSG00000113716), score: 0.6 HRH2histamine receptor H2 (ENSG00000113749), score: 0.56 HRH3histamine receptor H3 (ENSG00000101180), score: 0.6 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (ENSG00000102468), score: 0.71 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (ENSG00000005700), score: -0.48 IDEinsulin-degrading enzyme (ENSG00000119912), score: -0.51 ING3inhibitor of growth family, member 3 (ENSG00000071243), score: -0.49 INO80DINO80 complex subunit D (ENSG00000114933), score: -0.49 INTS8integrator complex subunit 8 (ENSG00000164941), score: -0.54 ITPK1inositol 1,3,4-triphosphate 5/6 kinase (ENSG00000100605), score: 0.62 ITPKAinositol 1,4,5-trisphosphate 3-kinase A (ENSG00000137825), score: 0.6 JAG2jagged 2 (ENSG00000184916), score: 0.53 KCNA6potassium voltage-gated channel, shaker-related subfamily, member 6 (ENSG00000151079), score: 0.55 KCNF1potassium voltage-gated channel, subfamily F, member 1 (ENSG00000162975), score: 0.75 KCNG1potassium voltage-gated channel, subfamily G, member 1 (ENSG00000026559), score: 0.69 KCNG3potassium voltage-gated channel, subfamily G, member 3 (ENSG00000171126), score: 0.67 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (ENSG00000135643), score: 0.58 KCNQ2potassium voltage-gated channel, KQT-like subfamily, member 2 (ENSG00000075043), score: 0.58 KCNQ5potassium voltage-gated channel, KQT-like subfamily, member 5 (ENSG00000185760), score: 0.6 KCNS1potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 (ENSG00000124134), score: 0.66 KCNS2potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 (ENSG00000156486), score: 0.57 KDM5Alysine (K)-specific demethylase 5A (ENSG00000073614), score: -0.5 KIAA0020KIAA0020 (ENSG00000080608), score: -0.51 KIAA0284KIAA0284 (ENSG00000099814), score: 0.63 KIAA0427KIAA0427 (ENSG00000134030), score: 0.55 KIAA1033KIAA1033 (ENSG00000136051), score: -0.55 KIAA1429KIAA1429 (ENSG00000164944), score: -0.54 KIF21Bkinesin family member 21B (ENSG00000116852), score: 0.55 KIRREL3kin of IRRE like 3 (Drosophila) (ENSG00000149571), score: 0.58 KLHDC3kelch domain containing 3 (ENSG00000124702), score: 0.6 KLHL22kelch-like 22 (Drosophila) (ENSG00000099910), score: 0.6 KLHL26kelch-like 26 (Drosophila) (ENSG00000167487), score: 0.64 LAPTM4Alysosomal protein transmembrane 4 alpha (ENSG00000068697), score: -0.55 LBRlamin B receptor (ENSG00000143815), score: -0.58 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: -0.53 LIMK1LIM domain kinase 1 (ENSG00000106683), score: 0.6 LMO4LIM domain only 4 (ENSG00000143013), score: 0.64 LMTK2lemur tyrosine kinase 2 (ENSG00000164715), score: 0.59 LOC100132308solute carrier family 29 (nucleoside transporters), member 4 pseudogene (ENSG00000164638), score: 0.58 LPPR3lipid phosphate phosphatase-related protein type 3 (ENSG00000129951), score: 0.73 LPPR4lipid phosphate phosphatase-related protein type 4 (ENSG00000117600), score: 0.57 LRFN2leucine rich repeat and fibronectin type III domain containing 2 (ENSG00000156564), score: 0.72 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (ENSG00000126243), score: 0.58 LRP3low density lipoprotein receptor-related protein 3 (ENSG00000130881), score: 0.6 LRTM2leucine-rich repeats and transmembrane domains 2 (ENSG00000166159), score: 0.61 LTV1LTV1 homolog (S. cerevisiae) (ENSG00000135521), score: -0.55 LYRM5LYR motif containing 5 (ENSG00000205707), score: -0.48 LYSMD3LysM, putative peptidoglycan-binding, domain containing 3 (ENSG00000176018), score: -0.54 LZICleucine zipper and CTNNBIP1 domain containing (ENSG00000162441), score: -0.61 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (ENSG00000197063), score: 0.56 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: -0.66 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (ENSG00000138834), score: 0.58 MARCH7membrane-associated ring finger (C3HC4) 7 (ENSG00000136536), score: -0.58 MATKmegakaryocyte-associated tyrosine kinase (ENSG00000007264), score: 0.66 MCEEmethylmalonyl CoA epimerase (ENSG00000124370), score: -0.5 MDM4Mdm4 p53 binding protein homolog (mouse) (ENSG00000198625), score: -0.62 MED7mediator complex subunit 7 (ENSG00000155868), score: -0.52 MEF2Cmyocyte enhancer factor 2C (ENSG00000081189), score: 0.55 MGAT5Bmannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B (ENSG00000167889), score: 0.63 MKL1megakaryoblastic leukemia (translocation) 1 (ENSG00000196588), score: 0.66 MKL2MKL/myocardin-like 2 (ENSG00000186260), score: 0.57 MKLN1muskelin 1, intracellular mediator containing kelch motifs (ENSG00000128585), score: -0.47 MRPL50mitochondrial ribosomal protein L50 (ENSG00000136897), score: -0.52 MTFR1mitochondrial fission regulator 1 (ENSG00000066855), score: -0.47 MTPAPmitochondrial poly(A) polymerase (ENSG00000107951), score: -0.65 MYNNmyoneurin (ENSG00000085274), score: -0.55 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (ENSG00000103174), score: 0.67 NCDNneurochondrin (ENSG00000020129), score: 0.61 NEDD1neural precursor cell expressed, developmentally down-regulated 1 (ENSG00000139350), score: -0.52 NELFnasal embryonic LHRH factor (ENSG00000165802), score: 0.56 NETO1neuropilin (NRP) and tolloid (TLL)-like 1 (ENSG00000166342), score: 0.56 NGEFneuronal guanine nucleotide exchange factor (ENSG00000066248), score: 0.56 NHLRC2NHL repeat containing 2 (ENSG00000196865), score: -0.52 NPTNneuroplastin (ENSG00000156642), score: 0.55 NTSR1neurotensin receptor 1 (high affinity) (ENSG00000101188), score: 0.62 NUDCD3NudC domain containing 3 (ENSG00000015676), score: 0.56 NUP54nucleoporin 54kDa (ENSG00000138750), score: -0.47 OCA2oculocutaneous albinism II (ENSG00000104044), score: 0.69 OLFM1olfactomedin 1 (ENSG00000130558), score: 0.55 OPN4opsin 4 (ENSG00000122375), score: 0.73 OPRL1opiate receptor-like 1 (ENSG00000125510), score: 0.63 OTOSotospiralin (ENSG00000178602), score: 0.55 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (ENSG00000122884), score: -0.46 PALMparalemmin (ENSG00000099864), score: 0.56 PAN3PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (ENSG00000152520), score: -0.47 PCDH19protocadherin 19 (ENSG00000165194), score: 0.58 PCSK1proprotein convertase subtilisin/kexin type 1 (ENSG00000175426), score: 0.56 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.55 PDE8Bphosphodiesterase 8B (ENSG00000113231), score: 0.6 PDZD7PDZ domain containing 7 (ENSG00000186862), score: 0.58 PEX13peroxisomal biogenesis factor 13 (ENSG00000162928), score: -0.57 PIGBphosphatidylinositol glycan anchor biosynthesis, class B (ENSG00000069943), score: -0.5 PIGQphosphatidylinositol glycan anchor biosynthesis, class Q (ENSG00000007541), score: 0.55 PIK3C2Aphosphoinositide-3-kinase, class 2, alpha polypeptide (ENSG00000011405), score: -0.53 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (ENSG00000105647), score: 0.61 PKP1plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) (ENSG00000081277), score: 0.59 PLEKHM2pleckstrin homology domain containing, family M (with RUN domain) member 2 (ENSG00000116786), score: 0.63 POLD3polymerase (DNA-directed), delta 3, accessory subunit (ENSG00000077514), score: -0.5 POLR3Apolymerase (RNA) III (DNA directed) polypeptide A, 155kDa (ENSG00000148606), score: 0.62 PPARDperoxisome proliferator-activated receptor delta (ENSG00000112033), score: 0.57 PPME1protein phosphatase methylesterase 1 (ENSG00000214517), score: 0.56 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.57 PRKAA1protein kinase, AMP-activated, alpha 1 catalytic subunit (ENSG00000132356), score: -0.46 PRKAR1Bprotein kinase, cAMP-dependent, regulatory, type I, beta (ENSG00000188191), score: 0.58 PRPF19PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae) (ENSG00000110107), score: 0.58 PRPF40APRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) (ENSG00000196504), score: -0.5 PSDpleckstrin and Sec7 domain containing (ENSG00000059915), score: 0.61 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: -0.49 PTHLHparathyroid hormone-like hormone (ENSG00000087494), score: 0.61 PTPRAprotein tyrosine phosphatase, receptor type, A (ENSG00000132670), score: 0.56 RAB40CRAB40C, member RAS oncogene family (ENSG00000197562), score: 0.81 RAI1retinoic acid induced 1 (ENSG00000108557), score: 0.55 RANGAP1Ran GTPase activating protein 1 (ENSG00000100401), score: 0.62 RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.66 RASD1RAS, dexamethasone-induced 1 (ENSG00000108551), score: 0.54 RBP3retinol binding protein 3, interstitial (ENSG00000107618), score: 0.62 RHOrhodopsin (ENSG00000163914), score: 0.54 RIMS3regulating synaptic membrane exocytosis 3 (ENSG00000117016), score: 0.56 RIOK2RIO kinase 2 (yeast) (ENSG00000058729), score: -0.56 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (ENSG00000067900), score: -0.71 RP2retinitis pigmentosa 2 (X-linked recessive) (ENSG00000102218), score: -0.51 RPF1ribosome production factor 1 homolog (S. cerevisiae) (ENSG00000117133), score: -0.5 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (ENSG00000067533), score: -0.47 RSPO2R-spondin 2 homolog (Xenopus laevis) (ENSG00000147655), score: 0.54 RTN4RL1reticulon 4 receptor-like 1 (ENSG00000185924), score: 0.58 SARSseryl-tRNA synthetase (ENSG00000031698), score: 0.57 SATB2SATB homeobox 2 (ENSG00000119042), score: 0.54 SCN3Bsodium channel, voltage-gated, type III, beta (ENSG00000166257), score: 0.58 SDC2syndecan 2 (ENSG00000169439), score: -0.46 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.52 SGSM2small G protein signaling modulator 2 (ENSG00000141258), score: 0.58 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: 0.62 SIN3BSIN3 homolog B, transcription regulator (yeast) (ENSG00000127511), score: 0.63 SIRT1sirtuin 1 (ENSG00000096717), score: -0.5 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (ENSG00000160326), score: 0.66 SLC6A7solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 (ENSG00000011083), score: 0.58 SMAD5SMAD family member 5 (ENSG00000113658), score: -0.64 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (ENSG00000100796), score: -0.53 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: -0.6 SNRNP48small nuclear ribonucleoprotein 48kDa (U11/U12) (ENSG00000168566), score: -0.46 SOX8SRY (sex determining region Y)-box 8 (ENSG00000005513), score: 0.54 SPDEFSAM pointed domain containing ets transcription factor (ENSG00000124664), score: 0.7 SREBF2sterol regulatory element binding transcription factor 2 (ENSG00000198911), score: 0.63 SSTsomatostatin (ENSG00000157005), score: 0.62 ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (ENSG00000117069), score: 0.61 STOML1stomatin (EPB72)-like 1 (ENSG00000067221), score: 0.66 STRN3striatin, calmodulin binding protein 3 (ENSG00000196792), score: -0.49 SYF2SYF2 homolog, RNA splicing factor (S. cerevisiae) (ENSG00000117614), score: -0.51 SYNGR3synaptogyrin 3 (ENSG00000127561), score: 0.55 SYT17synaptotagmin XVII (ENSG00000103528), score: 0.63 TAF12TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa (ENSG00000120656), score: -0.49 TAF1ATATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa (ENSG00000143498), score: -0.5 TBC1D23TBC1 domain family, member 23 (ENSG00000036054), score: -0.61 TBL3transducin (beta)-like 3 (ENSG00000183751), score: 0.6 TDRD3tudor domain containing 3 (ENSG00000083544), score: -0.67 TECPR1tectonin beta-propeller repeat containing 1 (ENSG00000205356), score: 0.53 TGFBR1transforming growth factor, beta receptor 1 (ENSG00000106799), score: -0.62 THOC2THO complex 2 (ENSG00000125676), score: -0.61 TMEM175transmembrane protein 175 (ENSG00000127419), score: 0.69 TMEM179transmembrane protein 179 (ENSG00000189203), score: 0.63 TMEM184Btransmembrane protein 184B (ENSG00000198792), score: 0.57 TMEM59Ltransmembrane protein 59-like (ENSG00000105696), score: 0.57 TOXthymocyte selection-associated high mobility group box (ENSG00000198846), score: 0.59 TP53INP1tumor protein p53 inducible nuclear protein 1 (ENSG00000164938), score: -0.5 TPPPtubulin polymerization promoting protein (ENSG00000171368), score: 0.58 TRAF7TNF receptor-associated factor 7 (ENSG00000131653), score: 0.57 TRIP4thyroid hormone receptor interactor 4 (ENSG00000103671), score: -0.47 TRMT61AtRNA methyltransferase 61 homolog A (S. cerevisiae) (ENSG00000166166), score: 0.63 TRPM2transient receptor potential cation channel, subfamily M, member 2 (ENSG00000142185), score: 0.62 TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: -0.66 TTC27tetratricopeptide repeat domain 27 (ENSG00000018699), score: -0.53 TUSC2tumor suppressor candidate 2 (ENSG00000114383), score: 0.57 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: -0.51 UNC5Aunc-5 homolog A (C. elegans) (ENSG00000113763), score: 0.63 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: -0.52 USP3ubiquitin specific peptidase 3 (ENSG00000140455), score: -0.53 USP8ubiquitin specific peptidase 8 (ENSG00000138592), score: -0.51 UTP11LUTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) (ENSG00000183520), score: -0.59 VPS11vacuolar protein sorting 11 homolog (S. cerevisiae) (ENSG00000160695), score: 0.55 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: -0.5 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: -0.55 WDR36WD repeat domain 36 (ENSG00000134987), score: -0.46 WDR37WD repeat domain 37 (ENSG00000047056), score: 0.62 WDR43WD repeat domain 43 (ENSG00000163811), score: -0.46 WEE1WEE1 homolog (S. pombe) (ENSG00000166483), score: -0.51 WNT7Awingless-type MMTV integration site family, member 7A (ENSG00000154764), score: 0.57 XRN15'-3' exoribonuclease 1 (ENSG00000114127), score: -0.58 XRN25'-3' exoribonuclease 2 (ENSG00000088930), score: -0.49 XYLT1xylosyltransferase I (ENSG00000103489), score: 0.58 YARStyrosyl-tRNA synthetase (ENSG00000134684), score: 0.56 YME1L1YME1-like 1 (S. cerevisiae) (ENSG00000136758), score: -0.5 YWHAHtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide (ENSG00000128245), score: 0.56 ZDHHC22zinc finger, DHHC-type containing 22 (ENSG00000177108), score: 0.62 ZER1zer-1 homolog (C. elegans) (ENSG00000160445), score: 0.62 ZFAND6zinc finger, AN1-type domain 6 (ENSG00000086666), score: -0.56
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppy_br_m_ca1 | ppy | br | m | _ |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |