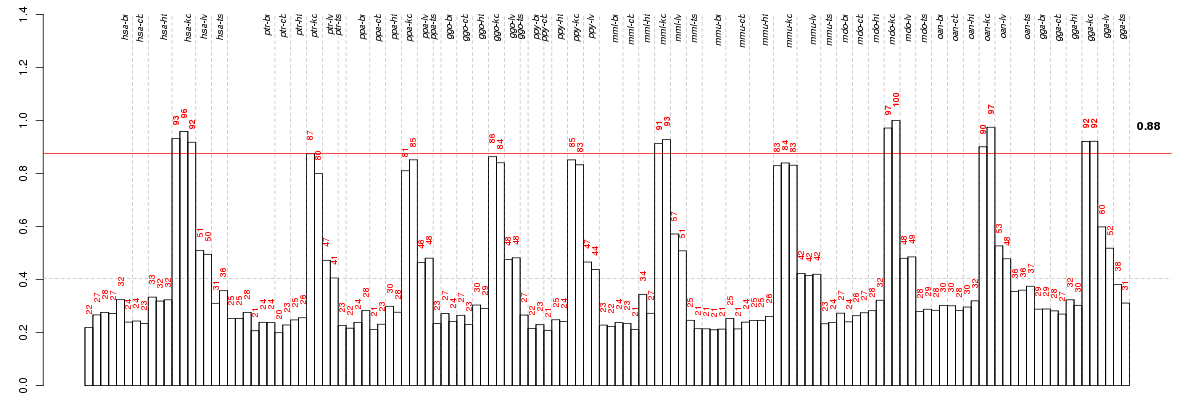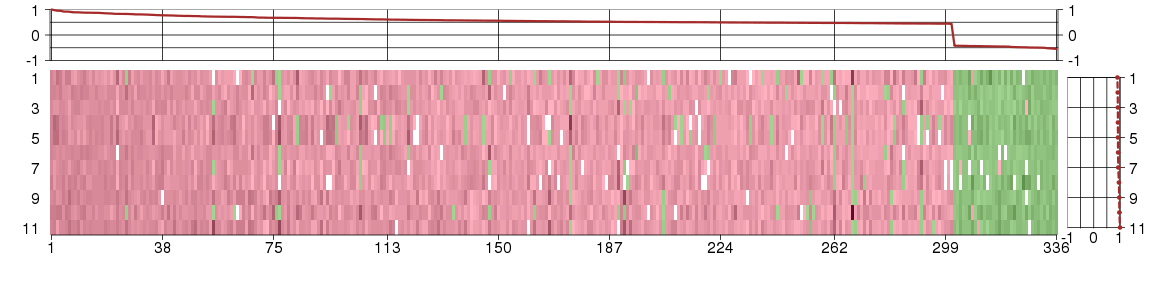

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
secretion
The controlled release of a substance by a cell, a group of cells, or a tissue.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cation transport
The directed movement of cations, atoms or small molecules with a net positive charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
sodium ion transport
The directed movement of sodium ions (Na+) into, out of, within or between cells by means of some external agent such as a transporter or pore.
metal ion transport
The directed movement of metal ions, any metal ion with an electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
embryo development
The process whose specific outcome is the progression of an embryo from its formation until the end of its embryonic life stage. The end of the embryonic stage is organism-specific. For example, for mammals, the process would begin with zygote formation and end with birth. For insects, the process would begin at zygote formation and end with larval hatching. For plant zygotic embryos, this would be from zygote formation to the end of seed dormancy. For plant vegetative embryos, this would be from the initial determination of the cell or group of cells to form an embryo until the point when the embryo becomes independent of the parent plant.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
monovalent inorganic cation transport
The directed movement of inorganic cations with a valency of one into, out of, within or between cells by means of some external agent such as a transporter or pore. Inorganic cations are atoms or small molecules with a positive charge which do not contain carbon in covalent linkage.
organic acid transport
The directed movement of organic acids, any acidic compound containing carbon in covalent linkage, into, out of, within or between cells by means of some external agent such as a transporter or pore.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
epithelial cell differentiation
The process whereby a relatively unspecialized cell acquires specialized features of an epithelial cell, any of the cells making up an epithelium.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
monocarboxylic acid metabolic process
The chemical reactions and pathways involving monocarboxylic acids, any organic acid containing one carboxyl (COOH) group or anion (COO-).
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
carboxylic acid transport
The directed movement of carboxylic acids into, out of, within or between cells by means of some external agent such as a transporter or pore. Carboxylic acids are organic acids containing one or more carboxyl (COOH) groups or anions (COO-).
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ development
Development, taking place during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic morphogenesis
The process by which anatomical structures are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form. The embryonic phase begins with zygote formation. The end of the embryonic phase is organism-specific. For example, it would be at birth for mammals, larval hatching for insects and seed dormancy in plants.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
epithelium development
The process whose specific outcome is the progression of an epithelium over time, from its formation to the mature structure. An epithelium is a tissue that covers the internal or external surfaces of an anatomical structure.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
embryo development
The process whose specific outcome is the progression of an embryo from its formation until the end of its embryonic life stage. The end of the embryonic stage is organism-specific. For example, for mammals, the process would begin with zygote formation and end with birth. For insects, the process would begin at zygote formation and end with larval hatching. For plant zygotic embryos, this would be from zygote formation to the end of seed dormancy. For plant vegetative embryos, this would be from the initial determination of the cell or group of cells to form an embryo until the point when the embryo becomes independent of the parent plant.
embryonic morphogenesis
The process by which anatomical structures are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form. The embryonic phase begins with zygote formation. The end of the embryonic phase is organism-specific. For example, it would be at birth for mammals, larval hatching for insects and seed dormancy in plants.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
embryonic organ development
Development, taking place during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
epithelial cell differentiation
The process whereby a relatively unspecialized cell acquires specialized features of an epithelial cell, any of the cells making up an epithelium.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
sodium ion transport
The directed movement of sodium ions (Na+) into, out of, within or between cells by means of some external agent such as a transporter or pore.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity. An example of this component is found in Mus musculus.
basement membrane
A thin layer of dense material found in various animal tissues interposed between the cells and the adjacent connective tissue. It consists of the basal lamina plus an associated layer of reticulin fibers.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
brush border
Dense covering of microvilli on the apical surface of epithelial cells in tissues such as the intestine, kidney, and choroid plexus; the microvilli aid absorption by increasing the surface area of the cell.
basolateral plasma membrane
The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
sodium channel complex
An ion channel complex through which sodium ions pass.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
apical part of cell
The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
basement membrane
A thin layer of dense material found in various animal tissues interposed between the cells and the adjacent connective tissue. It consists of the basal lamina plus an associated layer of reticulin fibers.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
peptidase activity
Catalysis of the hydrolysis of a peptide bond. A peptide bond is a covalent bond formed when the carbon atom from the carboxyl group of one amino acid shares electrons with the nitrogen atom from the amino group of a second amino acid.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
organic acid transmembrane transporter activity
Catalysis of the transfer of organic acids, any acidic compound containing carbon in covalent linkage, from one side of the membrane to the other.
secondary active transmembrane transporter activity
Catalysis of the transfer of a solute from one side of a membrane to the other, up its concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction and is driven by a chemiosmotic source of energy. Chemiosmotic sources of energy include uniport, symport or antiport.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
exopeptidase activity
Catalysis of the hydrolysis of a peptide bond not more than three residues from the N- or C-terminus of a polypeptide chain, in a reaction that requires a free N-terminal amino group, C-terminal carboxyl group or both.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
peptidase activity, acting on L-amino acid peptides
Catalysis of the hydrolysis of peptide bonds formed between L-amino acids.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
monovalent inorganic cation transmembrane transporter activity
Catalysis of the transfer of a inorganic cations with a valency of one from one side of a membrane to the other. Inorganic cations are atoms or small molecules with a positive charge that do not contain carbon in covalent linkage.
symporter activity
Enables the active transport of a solute across a membrane by a mechanism whereby two or more species are transported together in the same direction in a tightly coupled process not directly linked to a form of energy other than chemiosmotic energy.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
vitamin binding
Interacting selectively and non-covalently with a vitamin, one of a number of unrelated organic substances that occur in many foods in small amounts and that are necessary in trace amounts for the normal metabolic functioning of the body.
active transmembrane transporter activity
Catalysis of the transfer of a specific substance or related group of substances from one side of a membrane to the other, up the solute's concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction.
inorganic cation transmembrane transporter activity
Catalysis of the transfer of inorganic cations from one side of a membrane to the other. Inorganic cations are atoms or small molecules with a positive charge that do not contain carbon in covalent linkage.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
carboxylic acid transmembrane transporter activity
Catalysis of the transfer of carboxylic acids from one side of the membrane to the other. Carboxylic acids are organic acids containing one or more carboxyl (COOH) groups or anions (COO-).
cofactor binding
Interacting selectively and non-covalently with a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
coenzyme binding
Interacting selectively and non-covalently with a coenzyme, any of various nonprotein organic cofactors that are required, in addition to an enzyme and a substrate, for an enzymatic reaction to proceed.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04964 | 3.396e-04 | 0.5597 | 6 | 8 | Proximal tubule bicarbonate reclamation |
| 04142 | 2.139e-03 | 3.568 | 13 | 51 | Lysosome |
| 04960 | 3.590e-03 | 1.119 | 7 | 16 | Aldosterone-regulated sodium reabsorption |
| 04512 | 2.095e-02 | 2.449 | 9 | 35 | ECM-receptor interaction |
| 00380 | 2.150e-02 | 1.119 | 6 | 16 | Tryptophan metabolism |
| 00260 | 4.459e-02 | 0.9095 | 5 | 13 | Glycine, serine and threonine metabolism |
A2BP1ataxin 2-binding protein 1 (ENSG00000078328), score: -0.43 A2LD1AIG2-like domain 1 (ENSG00000134864), score: 0.51 ABCC4ATP-binding cassette, sub-family C (CFTR/MRP), member 4 (ENSG00000125257), score: 0.63 ACAD8acyl-CoA dehydrogenase family, member 8 (ENSG00000151498), score: 0.51 ACE2angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 (ENSG00000130234), score: 0.48 ACMSDaminocarboxymuconate semialdehyde decarboxylase (ENSG00000153086), score: 0.6 ACO1aconitase 1, soluble (ENSG00000122729), score: 0.51 ACOT11acyl-CoA thioesterase 11 (ENSG00000162390), score: 0.47 ACSM3acyl-CoA synthetase medium-chain family member 3 (ENSG00000005187), score: 0.59 ACSM5acyl-CoA synthetase medium-chain family member 5 (ENSG00000183549), score: 0.45 ADAM19ADAM metallopeptidase domain 19 (ENSG00000135074), score: -0.48 AGPHD1aminoglycoside phosphotransferase domain containing 1 (ENSG00000188266), score: 0.55 AGR2anterior gradient homolog 2 (Xenopus laevis) (ENSG00000106541), score: 0.62 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.47 AKAP6A kinase (PRKA) anchor protein 6 (ENSG00000151320), score: -0.48 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (ENSG00000117448), score: 0.55 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (ENSG00000119711), score: 0.55 ALDH8A1aldehyde dehydrogenase 8 family, member A1 (ENSG00000118514), score: 0.46 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.46 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.45 ANKRD2ankyrin repeat domain 2 (stretch responsive muscle) (ENSG00000165887), score: 0.46 ANKRD22ankyrin repeat domain 22 (ENSG00000152766), score: 0.54 ANKS4Bankyrin repeat and sterile alpha motif domain containing 4B (ENSG00000175311), score: 0.66 AP1G1adaptor-related protein complex 1, gamma 1 subunit (ENSG00000166747), score: 0.47 AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.54 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: 0.5 APLP2amyloid beta (A4) precursor-like protein 2 (ENSG00000084234), score: 0.58 AQP3aquaporin 3 (Gill blood group) (ENSG00000165272), score: 0.73 ARHGAP24Rho GTPase activating protein 24 (ENSG00000138639), score: 0.47 ARHGAP6Rho GTPase activating protein 6 (ENSG00000047648), score: 0.52 ARSBarylsulfatase B (ENSG00000113273), score: 0.52 ASAH1N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ENSG00000104763), score: 0.67 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ENSG00000153317), score: -0.49 ASPAaspartoacylase (ENSG00000108381), score: 0.65 ATP1B1ATPase, Na+/K+ transporting, beta 1 polypeptide (ENSG00000143153), score: 0.49 ATP6V0A4ATPase, H+ transporting, lysosomal V0 subunit a4 (ENSG00000105929), score: 0.87 ATP6V0D2ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 (ENSG00000147614), score: 0.76 ATP6V1C2ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 (ENSG00000143882), score: 0.48 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.51 BACE2beta-site APP-cleaving enzyme 2 (ENSG00000182240), score: 0.48 BAMBIBMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (ENSG00000095739), score: 0.49 BDKRB1bradykinin receptor B1 (ENSG00000100739), score: 0.56 BNC2basonuclin 2 (ENSG00000173068), score: 0.49 BPHLbiphenyl hydrolase-like (serine hydrolase) (ENSG00000137274), score: 0.49 BSPRYB-box and SPRY domain containing (ENSG00000119411), score: 0.51 C11orf54chromosome 11 open reading frame 54 (ENSG00000182919), score: 0.6 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.71 C2orf18chromosome 2 open reading frame 18 (ENSG00000213699), score: 0.61 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.71 C4orf31chromosome 4 open reading frame 31 (ENSG00000173376), score: 0.6 C9orf72chromosome 9 open reading frame 72 (ENSG00000147894), score: -0.49 CA12carbonic anhydrase XII (ENSG00000074410), score: 0.61 CALB1calbindin 1, 28kDa (ENSG00000104327), score: 0.47 CALML4calmodulin-like 4 (ENSG00000129007), score: 0.56 CAMK2Dcalcium/calmodulin-dependent protein kinase II delta (ENSG00000145349), score: -0.43 CAPN13calpain 13 (ENSG00000162949), score: 0.46 CASRcalcium-sensing receptor (ENSG00000036828), score: 0.88 CASS4Cas scaffolding protein family member 4 (ENSG00000087589), score: 0.51 CBR4carbonyl reductase 4 (ENSG00000145439), score: 0.45 CCDC88Acoiled-coil domain containing 88A (ENSG00000115355), score: -0.44 CDCP1CUB domain containing protein 1 (ENSG00000163814), score: 0.53 CDH1cadherin 1, type 1, E-cadherin (epithelial) (ENSG00000039068), score: 0.52 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (ENSG00000113361), score: 0.54 CELF2CUGBP, Elav-like family member 2 (ENSG00000048740), score: -0.55 CLCN5chloride channel 5 (ENSG00000171365), score: 0.69 CLDN16claudin 16 (ENSG00000113946), score: 0.88 CLDN19claudin 19 (ENSG00000164007), score: 0.91 CLIC6chloride intracellular channel 6 (ENSG00000159212), score: 0.83 CLN5ceroid-lipofuscinosis, neuronal 5 (ENSG00000102805), score: 0.45 CLN6ceroid-lipofuscinosis, neuronal 6, late infantile, variant (ENSG00000128973), score: 0.47 CLVS1clavesin 1 (ENSG00000177182), score: -0.42 CNNM2cyclin M2 (ENSG00000148842), score: 0.66 COL4A3collagen, type IV, alpha 3 (Goodpasture antigen) (ENSG00000169031), score: 0.71 COL4A4collagen, type IV, alpha 4 (ENSG00000081052), score: 0.7 CP110CP110 protein (ENSG00000103540), score: -0.45 CRYL1crystallin, lambda 1 (ENSG00000165475), score: 0.5 CTDSPLCTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like (ENSG00000144677), score: 0.61 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.44 CTSHcathepsin H (ENSG00000103811), score: 0.49 CUBNcubilin (intrinsic factor-cobalamin receptor) (ENSG00000107611), score: 0.95 CYBASC3cytochrome b, ascorbate dependent 3 (ENSG00000162144), score: 0.51 DAB2disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) (ENSG00000153071), score: 0.79 DAOD-amino-acid oxidase (ENSG00000110887), score: 0.65 DCUN1D5DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) (ENSG00000137692), score: -0.47 DDCdopa decarboxylase (aromatic L-amino acid decarboxylase) (ENSG00000132437), score: 0.57 DENND2DDENN/MADD domain containing 2D (ENSG00000162777), score: 0.64 DENND5ADENN/MADD domain containing 5A (ENSG00000184014), score: -0.44 DHDPSLdihydrodipicolinate synthase-like, mitochondrial (ENSG00000241935), score: 0.67 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.48 DMGDHdimethylglycine dehydrogenase (ENSG00000132837), score: 0.51 DOCK4dedicator of cytokinesis 4 (ENSG00000128512), score: -0.45 DPEP1dipeptidase 1 (renal) (ENSG00000015413), score: 0.78 DPP4dipeptidyl-peptidase 4 (ENSG00000197635), score: 0.73 DPYSdihydropyrimidinase (ENSG00000147647), score: 0.56 EFEMP1EGF-containing fibulin-like extracellular matrix protein 1 (ENSG00000115380), score: 0.5 EGFepidermal growth factor (ENSG00000138798), score: 0.57 EHFets homologous factor (ENSG00000135373), score: 0.75 EHHADHenoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (ENSG00000113790), score: 0.47 ELF3E74-like factor 3 (ets domain transcription factor, epithelial-specific ) (ENSG00000163435), score: 0.67 ELF5E74-like factor 5 (ets domain transcription factor) (ENSG00000135374), score: 0.83 ELMO3engulfment and cell motility 3 (ENSG00000102890), score: 0.52 ENPEPglutamyl aminopeptidase (aminopeptidase A) (ENSG00000138792), score: 0.61 EPCAMepithelial cell adhesion molecule (ENSG00000119888), score: 0.7 ERBB2v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) (ENSG00000141736), score: 0.5 EREGepiregulin (ENSG00000124882), score: 0.53 ESRP1epithelial splicing regulatory protein 1 (ENSG00000104413), score: 0.75 EVCEllis van Creveld syndrome (ENSG00000072840), score: 0.59 F2RL1coagulation factor II (thrombin) receptor-like 1 (ENSG00000164251), score: 0.77 FAAHfatty acid amide hydrolase (ENSG00000117480), score: 0.49 FAM160B1family with sequence similarity 160, member B1 (ENSG00000151553), score: -0.47 FAM167Bfamily with sequence similarity 167, member B (ENSG00000183615), score: 0.44 FAM3Dfamily with sequence similarity 3, member D (ENSG00000198643), score: 0.68 FAM83Bfamily with sequence similarity 83, member B (ENSG00000168143), score: 0.67 FBXW7F-box and WD repeat domain containing 7 (ENSG00000109670), score: -0.5 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.5 FGF14fibroblast growth factor 14 (ENSG00000102466), score: -0.45 FGFR4fibroblast growth factor receptor 4 (ENSG00000160867), score: 0.49 FOXI1forkhead box I1 (ENSG00000168269), score: 0.89 FRAS1Fraser syndrome 1 (ENSG00000138759), score: 0.71 FSHBfollicle stimulating hormone, beta polypeptide (ENSG00000131808), score: 0.47 FZD4frizzled homolog 4 (Drosophila) (ENSG00000174804), score: 0.48 FZD6frizzled homolog 6 (Drosophila) (ENSG00000164930), score: 0.5 GALK2galactokinase 2 (ENSG00000156958), score: 0.62 GALNT11UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) (ENSG00000178234), score: 0.59 GALNT14UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) (ENSG00000158089), score: 0.74 GATA3GATA binding protein 3 (ENSG00000107485), score: 0.86 GCM1glial cells missing homolog 1 (Drosophila) (ENSG00000137270), score: 0.54 GLDCglycine dehydrogenase (decarboxylating) (ENSG00000178445), score: 0.48 GOPCgolgi-associated PDZ and coiled-coil motif containing (ENSG00000047932), score: -0.43 GPR114G protein-coupled receptor 114 (ENSG00000159618), score: 0.52 GPR160G protein-coupled receptor 160 (ENSG00000173890), score: 0.5 GPR56G protein-coupled receptor 56 (ENSG00000205336), score: 0.48 GPRC5CG protein-coupled receptor, family C, group 5, member C (ENSG00000170412), score: 0.48 GRAMD2GRAM domain containing 2 (ENSG00000175318), score: 0.46 GRHL2grainyhead-like 2 (Drosophila) (ENSG00000083307), score: 0.84 GSCgoosecoid homeobox (ENSG00000133937), score: 0.67 HAO2hydroxyacid oxidase 2 (long chain) (ENSG00000116882), score: 0.49 HDHD3haloacid dehalogenase-like hydrolase domain containing 3 (ENSG00000119431), score: 0.49 HGDhomogentisate 1,2-dioxygenase (ENSG00000113924), score: 0.46 HNF1AHNF1 homeobox A (ENSG00000135100), score: 0.57 HNF4Ahepatocyte nuclear factor 4, alpha (ENSG00000101076), score: 0.57 HNF4Ghepatocyte nuclear factor 4, gamma (ENSG00000164749), score: 0.68 HOXA10homeobox A10 (ENSG00000153807), score: 0.81 HOXB7homeobox B7 (ENSG00000120087), score: 0.8 HOXD10homeobox D10 (ENSG00000128710), score: 0.95 HOXD4homeobox D4 (ENSG00000170166), score: 0.87 HSD11B2hydroxysteroid (11-beta) dehydrogenase 2 (ENSG00000176387), score: 0.88 IGFBP7insulin-like growth factor binding protein 7 (ENSG00000163453), score: 0.46 ILDR1immunoglobulin-like domain containing receptor 1 (ENSG00000145103), score: 0.71 IRF6interferon regulatory factor 6 (ENSG00000117595), score: 0.52 IRX2iroquois homeobox 2 (ENSG00000170561), score: 0.57 ITGA8integrin, alpha 8 (ENSG00000077943), score: 0.45 ITGB4integrin, beta 4 (ENSG00000132470), score: 0.49 ITGB6integrin, beta 6 (ENSG00000115221), score: 0.51 IVDisovaleryl-CoA dehydrogenase (ENSG00000128928), score: 0.45 IVNS1ABPinfluenza virus NS1A binding protein (ENSG00000116679), score: 0.5 JAG1jagged 1 (ENSG00000101384), score: 0.52 KCNJ1potassium inwardly-rectifying channel, subfamily J, member 1 (ENSG00000151704), score: 0.83 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (ENSG00000153822), score: 0.75 KCNK5potassium channel, subfamily K, member 5 (ENSG00000164626), score: 0.67 KIAA1522KIAA1522 (ENSG00000162522), score: 0.59 KIF12kinesin family member 12 (ENSG00000136883), score: 0.56 KIF13Bkinesin family member 13B (ENSG00000197892), score: 0.5 KIRRELkin of IRRE like (Drosophila) (ENSG00000183853), score: 0.52 KLklotho (ENSG00000133116), score: 0.77 KLHDC7Akelch domain containing 7A (ENSG00000179023), score: 0.63 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.64 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (ENSG00000117009), score: 0.49 LAD1ladinin 1 (ENSG00000159166), score: 0.67 LAMA1laminin, alpha 1 (ENSG00000101680), score: 0.47 LAMB1laminin, beta 1 (ENSG00000091136), score: 0.49 LAMC3laminin, gamma 3 (ENSG00000050555), score: 0.51 LGALS2lectin, galactoside-binding, soluble, 2 (ENSG00000100079), score: 0.83 LGMNlegumain (ENSG00000100600), score: 0.77 LLGL2lethal giant larvae homolog 2 (Drosophila) (ENSG00000073350), score: 0.63 LMX1BLIM homeobox transcription factor 1, beta (ENSG00000136944), score: 0.77 LOC100290519similar to transcription factor MEL1 (ENSG00000142611), score: 0.5 LRRC58leucine rich repeat containing 58 (ENSG00000163428), score: 0.45 LYPD2LY6/PLAUR domain containing 2 (ENSG00000197353), score: 0.47 MALmal, T-cell differentiation protein (ENSG00000172005), score: 0.47 MARVELD2MARVEL domain containing 2 (ENSG00000152939), score: 0.5 MARVELD3MARVEL domain containing 3 (ENSG00000140832), score: 0.72 MBOAT1membrane bound O-acyltransferase domain containing 1 (ENSG00000172197), score: 0.45 MPP1membrane protein, palmitoylated 1, 55kDa (ENSG00000130830), score: 0.51 MPZL2myelin protein zero-like 2 (ENSG00000149573), score: 0.54 MSRAmethionine sulfoxide reductase A (ENSG00000175806), score: 0.5 MYH9myosin, heavy chain 9, non-muscle (ENSG00000100345), score: 0.5 MYO1Dmyosin ID (ENSG00000176658), score: 0.53 MYO6myosin VI (ENSG00000196586), score: 0.49 NEK8NIMA (never in mitosis gene a)- related kinase 8 (ENSG00000160602), score: 0.49 NHSNance-Horan syndrome (congenital cataracts and dental anomalies) (ENSG00000188158), score: 0.48 NIT2nitrilase family, member 2 (ENSG00000114021), score: 0.54 NME7non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000143156), score: 0.51 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.78 NPHS2nephrosis 2, idiopathic, steroid-resistant (podocin) (ENSG00000116218), score: 0.82 NPNTnephronectin (ENSG00000168743), score: 0.6 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (ENSG00000113389), score: 0.68 NRN1neuritin 1 (ENSG00000124785), score: -0.42 NTN4netrin 4 (ENSG00000074527), score: 0.53 NUAK2NUAK family, SNF1-like kinase, 2 (ENSG00000163545), score: 0.65 OSTalphaorganic solute transporter alpha (ENSG00000163959), score: 0.51 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.48 PAPPApregnancy-associated plasma protein A, pappalysin 1 (ENSG00000182752), score: 0.47 PAPPA2pappalysin 2 (ENSG00000116183), score: 0.45 PAQR5progestin and adipoQ receptor family member V (ENSG00000137819), score: 0.72 PCK1phosphoenolpyruvate carboxykinase 1 (soluble) (ENSG00000124253), score: 0.55 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.58 PDCD5programmed cell death 5 (ENSG00000105185), score: -0.48 PDE4Bphosphodiesterase 4B, cAMP-specific (ENSG00000184588), score: -0.44 PDS5BPDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) (ENSG00000083642), score: -0.45 PEPDpeptidase D (ENSG00000124299), score: 0.72 PGPEP1pyroglutamyl-peptidase I (ENSG00000130517), score: 0.57 PKD2polycystic kidney disease 2 (autosomal dominant) (ENSG00000118762), score: 0.53 PLA2G15phospholipase A2, group XV (ENSG00000103066), score: 0.49 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (ENSG00000197943), score: 0.55 PLEKHA7pleckstrin homology domain containing, family A member 7 (ENSG00000166689), score: 0.45 PLEKHF2pleckstrin homology domain containing, family F (with FYVE domain) member 2 (ENSG00000175895), score: 0.5 PLS1plastin 1 (ENSG00000120756), score: 0.63 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (ENSG00000111725), score: 0.48 PRLRprolactin receptor (ENSG00000113494), score: 0.58 PROM2prominin 2 (ENSG00000155066), score: 0.61 PRSS23protease, serine, 23 (ENSG00000150687), score: 0.48 PTERphosphotriesterase related (ENSG00000165983), score: 0.44 PTGER4prostaglandin E receptor 4 (subtype EP4) (ENSG00000171522), score: 0.56 PTPRQprotein tyrosine phosphatase, receptor type, Q (ENSG00000139304), score: 0.64 PYROXD2pyridine nucleotide-disulphide oxidoreductase domain 2 (ENSG00000119943), score: 0.67 RAB11FIP3RAB11 family interacting protein 3 (class II) (ENSG00000090565), score: 0.48 RAB17RAB17, member RAS oncogene family (ENSG00000124839), score: 0.45 RAB19RAB19, member RAS oncogene family (ENSG00000146955), score: 0.79 RAB20RAB20, member RAS oncogene family (ENSG00000139832), score: 0.46 RAB7L1RAB7, member RAS oncogene family-like 1 (ENSG00000117280), score: 0.65 RASA3RAS p21 protein activator 3 (ENSG00000185989), score: -0.5 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.64 RB1CC1RB1-inducible coiled-coil 1 (ENSG00000023287), score: -0.48 RBKSribokinase (ENSG00000171174), score: 0.51 REEP1receptor accessory protein 1 (ENSG00000068615), score: -0.53 RGNregucalcin (senescence marker protein-30) (ENSG00000130988), score: 0.45 RHAGRh-associated glycoprotein (ENSG00000112077), score: 0.63 RHCGRh family, C glycoprotein (ENSG00000140519), score: 0.87 RIPK4receptor-interacting serine-threonine kinase 4 (ENSG00000183421), score: 0.52 RNF152ring finger protein 152 (ENSG00000176641), score: 0.54 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (ENSG00000117676), score: 0.48 SACSspastic ataxia of Charlevoix-Saguenay (sacsin) (ENSG00000151835), score: -0.52 SALL1sal-like 1 (Drosophila) (ENSG00000103449), score: 0.54 SCAMP2secretory carrier membrane protein 2 (ENSG00000140497), score: 0.54 SCINscinderin (ENSG00000006747), score: 0.67 SCNN1Asodium channel, nonvoltage-gated 1 alpha (ENSG00000111319), score: 0.8 SCNN1Bsodium channel, nonvoltage-gated 1, beta (ENSG00000168447), score: 0.92 SCNN1Gsodium channel, nonvoltage-gated 1, gamma (ENSG00000166828), score: 0.85 SCRN2secernin 2 (ENSG00000141295), score: 0.52 SDK1sidekick homolog 1, cell adhesion molecule (chicken) (ENSG00000146555), score: 0.46 SDSLserine dehydratase-like (ENSG00000139410), score: 0.59 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (ENSG00000001617), score: 0.54 SFXN2sideroflexin 2 (ENSG00000156398), score: 0.68 SGK1serum/glucocorticoid regulated kinase 1 (ENSG00000118515), score: 0.5 SGK2serum/glucocorticoid regulated kinase 2 (ENSG00000101049), score: 0.51 SH2D4ASH2 domain containing 4A (ENSG00000104611), score: 0.48 SHISA2shisa homolog 2 (Xenopus laevis) (ENSG00000180730), score: 0.65 SIM1single-minded homolog 1 (Drosophila) (ENSG00000112246), score: 0.97 SIM2single-minded homolog 2 (Drosophila) (ENSG00000159263), score: 0.51 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.76 SLC13A1solute carrier family 13 (sodium/sulfate symporters), member 1 (ENSG00000081800), score: 0.71 SLC13A3solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 (ENSG00000158296), score: 0.75 SLC16A12solute carrier family 16, member 12 (monocarboxylic acid transporter 12) (ENSG00000152779), score: 0.68 SLC16A2solute carrier family 16, member 2 (monocarboxylic acid transporter 8) (ENSG00000147100), score: 0.5 SLC16A4solute carrier family 16, member 4 (monocarboxylic acid transporter 5) (ENSG00000168679), score: 0.87 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (ENSG00000119899), score: 0.46 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.51 SLC1A2solute carrier family 1 (glial high affinity glutamate transporter), member 2 (ENSG00000110436), score: -0.43 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.63 SLC22A7solute carrier family 22 (organic anion transporter), member 7 (ENSG00000137204), score: 0.55 SLC25A10solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 (ENSG00000183048), score: 0.52 SLC25A48solute carrier family 25, member 48 (ENSG00000145832), score: 0.63 SLC26A7solute carrier family 26, member 7 (ENSG00000147606), score: 0.6 SLC34A1solute carrier family 34 (sodium phosphate), member 1 (ENSG00000131183), score: 0.92 SLC35F2solute carrier family 35, member F2 (ENSG00000110660), score: 0.59 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (ENSG00000138079), score: 0.62 SLC44A3solute carrier family 44, member 3 (ENSG00000143036), score: 0.57 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (ENSG00000080493), score: 0.55 SLC5A11solute carrier family 5 (sodium/glucose cotransporter), member 11 (ENSG00000158865), score: 0.54 SLC5A12solute carrier family 5 (sodium/glucose cotransporter), member 12 (ENSG00000148942), score: 0.89 SLC5A9solute carrier family 5 (sodium/glucose cotransporter), member 9 (ENSG00000117834), score: 0.74 SLC6A19solute carrier family 6 (neutral amino acid transporter), member 19 (ENSG00000174358), score: 1 SLC7A9solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 (ENSG00000021488), score: 0.73 SLC9A3solute carrier family 9 (sodium/hydrogen exchanger), member 3 (ENSG00000066230), score: 0.59 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (ENSG00000109062), score: 0.56 SLC9A4solute carrier family 9 (sodium/hydrogen exchanger), member 4 (ENSG00000180251), score: 0.55 SMYD2SET and MYND domain containing 2 (ENSG00000143499), score: -0.42 SNX2sorting nexin 2 (ENSG00000205302), score: 0.46 SPP1secreted phosphoprotein 1 (ENSG00000118785), score: 0.55 SRMSsrc-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites (ENSG00000125508), score: 0.59 ST14suppression of tumorigenicity 14 (colon carcinoma) (ENSG00000149418), score: 0.72 STC1stanniocalcin 1 (ENSG00000159167), score: 0.59 SULF2sulfatase 2 (ENSG00000196562), score: 0.58 SUMF1sulfatase modifying factor 1 (ENSG00000144455), score: 0.44 SUSD2sushi domain containing 2 (ENSG00000099994), score: 0.85 SUSD3sushi domain containing 3 (ENSG00000157303), score: 0.63 TARSL2threonyl-tRNA synthetase-like 2 (ENSG00000185418), score: -0.43 TBX2T-box 2 (ENSG00000121068), score: 0.57 TCF21transcription factor 21 (ENSG00000118526), score: 0.48 TCN2transcobalamin II (ENSG00000185339), score: 0.6 TFAP2Atranscription factor AP-2 alpha (activating enhancer binding protein 2 alpha) (ENSG00000137203), score: 0.58 TFAP2Btranscription factor AP-2 beta (activating enhancer binding protein 2 beta) (ENSG00000008196), score: 0.66 TFCP2L1transcription factor CP2-like 1 (ENSG00000115112), score: 0.83 THBS1thrombospondin 1 (ENSG00000137801), score: 0.48 THNSL2threonine synthase-like 2 (S. cerevisiae) (ENSG00000144115), score: 0.52 TIMP3TIMP metallopeptidase inhibitor 3 (ENSG00000100234), score: 0.47 TINAGtubulointerstitial nephritis antigen (ENSG00000137251), score: 0.81 TMCC1transmembrane and coiled-coil domain family 1 (ENSG00000172765), score: -0.44 TMCO4transmembrane and coiled-coil domains 4 (ENSG00000162542), score: 0.45 TMEM164transmembrane protein 164 (ENSG00000157600), score: 0.48 TMEM171transmembrane protein 171 (ENSG00000157111), score: 0.81 TMEM27transmembrane protein 27 (ENSG00000147003), score: 0.82 TMEM51transmembrane protein 51 (ENSG00000171729), score: 0.57 TMEM82transmembrane protein 82 (ENSG00000162460), score: 0.45 TMIGD1transmembrane and immunoglobulin domain containing 1 (ENSG00000182271), score: 0.7 TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.72 TMPRSS2transmembrane protease, serine 2 (ENSG00000184012), score: 0.72 TNFSF15tumor necrosis factor (ligand) superfamily, member 15 (ENSG00000181634), score: 0.56 TOX3TOX high mobility group box family member 3 (ENSG00000103460), score: 0.52 TPP1tripeptidyl peptidase I (ENSG00000166340), score: 0.57 TRPM6transient receptor potential cation channel, subfamily M, member 6 (ENSG00000119121), score: 0.74 TRPV4transient receptor potential cation channel, subfamily V, member 4 (ENSG00000111199), score: 0.64 TSG101tumor susceptibility gene 101 (ENSG00000074319), score: 0.46 TTC36tetratricopeptide repeat domain 36 (ENSG00000172425), score: 0.44 TTLL7tubulin tyrosine ligase-like family, member 7 (ENSG00000137941), score: -0.5 UBA2ubiquitin-like modifier activating enzyme 2 (ENSG00000126261), score: -0.44 UBLCP1ubiquitin-like domain containing CTD phosphatase 1 (ENSG00000164332), score: -0.46 UPB1ureidopropionase, beta (ENSG00000100024), score: 0.55 WARS2tryptophanyl tRNA synthetase 2, mitochondrial (ENSG00000116874), score: 0.47 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.64 WNK1WNK lysine deficient protein kinase 1 (ENSG00000060237), score: 0.58 XPNPEP1X-prolyl aminopeptidase (aminopeptidase P) 1, soluble (ENSG00000108039), score: 0.48 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.89 ZEB1zinc finger E-box binding homeobox 1 (ENSG00000148516), score: -0.53 ZFPM2zinc finger protein, multitype 2 (ENSG00000169946), score: -0.5
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_kd_m_ca1 | oan | kd | m | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| gga_kd_m_ca1 | gga | kd | m | _ |
| gga_kd_f_ca1 | gga | kd | f | _ |
| mml_kd_f_ca1 | mml | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| oan_kd_f_ca1 | oan | kd | f | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |