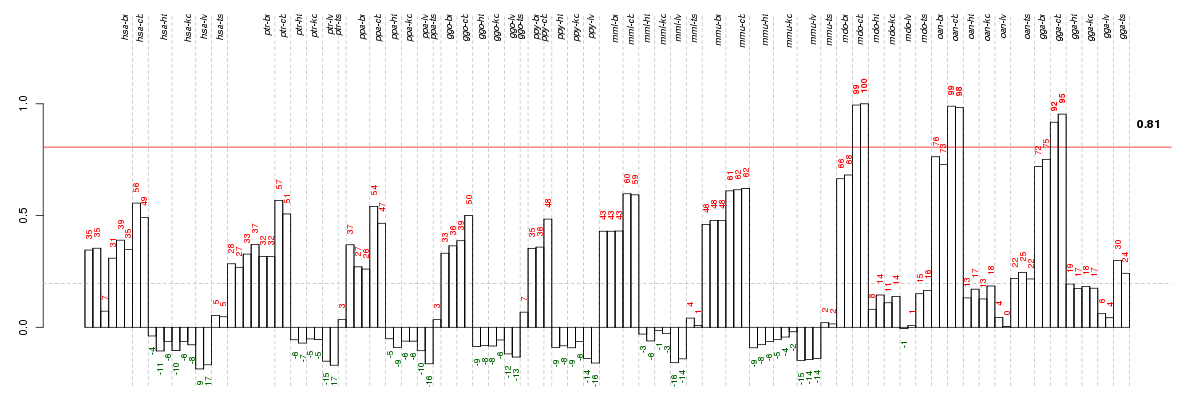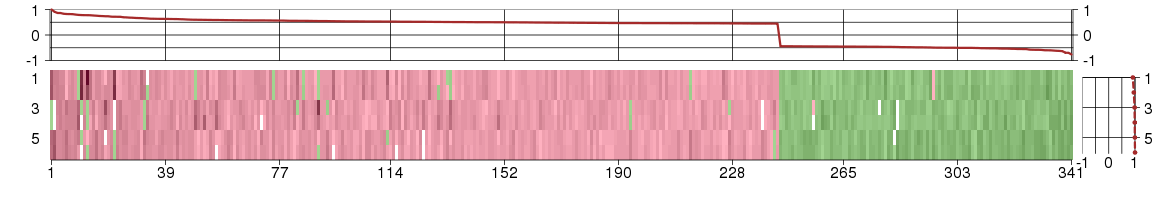

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
nerve-nerve synaptic transmission
The process of communication from a neuron to another neuron across a synapse.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
synaptic transmission, glutamatergic
The process of communication from a neuron to another neuron across a synapse using the neurotransmitter glutamate.
all
NA
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.


ABHD3abhydrolase domain containing 3 (ENSG00000158201), score: 0.57 ADAM22ADAM metallopeptidase domain 22 (ENSG00000008277), score: 0.55 ADAMTS2ADAM metallopeptidase with thrombospondin type 1 motif, 2 (ENSG00000087116), score: 0.46 ADARB2adenosine deaminase, RNA-specific, B2 (ENSG00000185736), score: 0.56 ADCYAP1adenylate cyclase activating polypeptide 1 (pituitary) (ENSG00000141433), score: 0.5 ADMadrenomedullin (ENSG00000148926), score: -0.44 AKAP1A kinase (PRKA) anchor protein 1 (ENSG00000121057), score: -0.46 ALG14asparagine-linked glycosylation 14 homolog (S. cerevisiae) (ENSG00000172339), score: -0.7 AMOTangiomotin (ENSG00000126016), score: 0.47 AMZ1archaelysin family metallopeptidase 1 (ENSG00000174945), score: 0.55 APPBP2amyloid beta precursor protein (cytoplasmic tail) binding protein 2 (ENSG00000062725), score: 0.46 ARPP21cAMP-regulated phosphoprotein, 21kDa (ENSG00000172995), score: 0.49 ARRDC4arrestin domain containing 4 (ENSG00000140450), score: -0.46 ASAMadipocyte-specific adhesion molecule (ENSG00000166250), score: 0.71 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.5 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (ENSG00000113732), score: -0.48 AURKAIP1aurora kinase A interacting protein 1 (ENSG00000175756), score: -0.5 AVPR1Barginine vasopressin receptor 1B (ENSG00000198049), score: 0.6 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (ENSG00000086062), score: -0.47 B4GALT6UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (ENSG00000118276), score: 0.46 BET3LBET3 like (S. cerevisiae) (ENSG00000173626), score: 0.72 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.61 C11orf41chromosome 11 open reading frame 41 (ENSG00000110427), score: 0.47 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.53 C13orf23chromosome 13 open reading frame 23 (ENSG00000120685), score: 0.55 C14orf37chromosome 14 open reading frame 37 (ENSG00000139971), score: 0.6 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.64 C15orf60chromosome 15 open reading frame 60 (ENSG00000183324), score: 0.53 C17orf75chromosome 17 open reading frame 75 (ENSG00000108666), score: 0.53 C18orf1chromosome 18 open reading frame 1 (ENSG00000168675), score: 0.53 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.63 C3orf17chromosome 3 open reading frame 17 (ENSG00000163608), score: -0.45 C3orf26chromosome 3 open reading frame 26 (ENSG00000184220), score: -0.46 C3orf58chromosome 3 open reading frame 58 (ENSG00000181744), score: 0.45 C3orf59chromosome 3 open reading frame 59 (ENSG00000180611), score: 0.47 C6orf106chromosome 6 open reading frame 106 (ENSG00000196821), score: -0.54 C6orf154chromosome 6 open reading frame 154 (ENSG00000204052), score: 0.5 C6orf162chromosome 6 open reading frame 162 (ENSG00000111850), score: -0.51 C6orf72chromosome 6 open reading frame 72 (ENSG00000055211), score: -0.59 C9orf4chromosome 9 open reading frame 4 (ENSG00000136805), score: 0.49 CA10carbonic anhydrase X (ENSG00000154975), score: 0.49 CA8carbonic anhydrase VIII (ENSG00000178538), score: 0.58 CACHD1cache domain containing 1 (ENSG00000158966), score: 0.59 CACNB4calcium channel, voltage-dependent, beta 4 subunit (ENSG00000182389), score: 0.47 CACNG2calcium channel, voltage-dependent, gamma subunit 2 (ENSG00000166862), score: 0.57 CALB1calbindin 1, 28kDa (ENSG00000104327), score: 0.47 CASD1CAS1 domain containing 1 (ENSG00000127995), score: 0.52 CASKcalcium/calmodulin-dependent serine protein kinase (MAGUK family) (ENSG00000147044), score: 0.47 CASP2caspase 2, apoptosis-related cysteine peptidase (ENSG00000106144), score: 0.52 CASP7caspase 7, apoptosis-related cysteine peptidase (ENSG00000165806), score: -0.53 CASTcalpastatin (ENSG00000153113), score: -0.57 CBFBcore-binding factor, beta subunit (ENSG00000067955), score: -0.45 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.75 CCBL2cysteine conjugate-beta lyase 2 (ENSG00000137944), score: -0.45 CCDC109Acoiled-coil domain containing 109A (ENSG00000156026), score: 0.53 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: -0.44 CCNYcyclin Y (ENSG00000108100), score: -0.5 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.46 CDH20cadherin 20, type 2 (ENSG00000101542), score: 0.49 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.48 CDK15cyclin-dependent kinase 15 (ENSG00000138395), score: 0.5 CDK17cyclin-dependent kinase 17 (ENSG00000059758), score: 0.58 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.62 CHD6chromodomain helicase DNA binding protein 6 (ENSG00000124177), score: 0.48 CHGBchromogranin B (secretogranin 1) (ENSG00000089199), score: 0.49 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: -0.54 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.47 CHRNB4cholinergic receptor, nicotinic, beta 4 (ENSG00000117971), score: 0.79 CINPcyclin-dependent kinase 2 interacting protein (ENSG00000100865), score: -0.55 CLCN3chloride channel 3 (ENSG00000109572), score: 0.46 CLPTM1LCLPTM1-like (ENSG00000049656), score: -0.55 CLVS2clavesin 2 (ENSG00000146352), score: 0.6 CNKSR2connector enhancer of kinase suppressor of Ras 2 (ENSG00000149970), score: 0.46 CNR1cannabinoid receptor 1 (brain) (ENSG00000118432), score: 0.64 CNTN4contactin 4 (ENSG00000144619), score: 0.53 CNTN5contactin 5 (ENSG00000149972), score: 0.56 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.51 CRB1crumbs homolog 1 (Drosophila) (ENSG00000134376), score: 0.55 CRHR1corticotropin releasing hormone receptor 1 (ENSG00000120088), score: 0.53 CRTAPcartilage associated protein (ENSG00000170275), score: -0.55 CRY1cryptochrome 1 (photolyase-like) (ENSG00000008405), score: 0.46 CTNND1catenin (cadherin-associated protein), delta 1 (ENSG00000198561), score: -0.49 CUL4Acullin 4A (ENSG00000139842), score: -0.6 CYR61cysteine-rich, angiogenic inducer, 61 (ENSG00000142871), score: -0.46 DCNdecorin (ENSG00000011465), score: -0.44 DCTDdCMP deaminase (ENSG00000129187), score: -0.52 DDX24DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 (ENSG00000089737), score: -0.45 DDX47DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (ENSG00000213782), score: -0.51 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.52 DNM3dynamin 3 (ENSG00000197959), score: 0.45 DNMT3ADNA (cytosine-5-)-methyltransferase 3 alpha (ENSG00000119772), score: 0.46 DOPEY2dopey family member 2 (ENSG00000142197), score: 0.47 DUSP10dual specificity phosphatase 10 (ENSG00000143507), score: 0.47 EGFL6EGF-like-domain, multiple 6 (ENSG00000198759), score: 0.54 EIF3Beukaryotic translation initiation factor 3, subunit B (ENSG00000106263), score: -0.55 EIF5Beukaryotic translation initiation factor 5B (ENSG00000158417), score: -0.5 ELAVL2ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) (ENSG00000107105), score: 0.47 ELAVL4ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) (ENSG00000162374), score: 0.45 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (ENSG00000118402), score: 0.46 EN1engrailed homeobox 1 (ENSG00000163064), score: 1 EPC1enhancer of polycomb homolog 1 (Drosophila) (ENSG00000120616), score: 0.46 EPGNepithelial mitogen homolog (mouse) (ENSG00000182585), score: 0.91 EPHA3EPH receptor A3 (ENSG00000044524), score: 0.58 EPHA8EPH receptor A8 (ENSG00000070886), score: 0.51 EPS15epidermal growth factor receptor pathway substrate 15 (ENSG00000085832), score: 0.52 ERI1exoribonuclease 1 (ENSG00000104626), score: 0.46 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.46 ESR2estrogen receptor 2 (ER beta) (ENSG00000140009), score: 0.51 FADDFas (TNFRSF6)-associated via death domain (ENSG00000168040), score: -0.45 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.45 FAM53Afamily with sequence similarity 53, member A (ENSG00000174137), score: 0.46 FAM69Afamily with sequence similarity 69, member A (ENSG00000154511), score: 0.51 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: -0.46 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.8 FBXO22F-box protein 22 (ENSG00000167196), score: 0.62 FGF14fibroblast growth factor 14 (ENSG00000102466), score: 0.51 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.76 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.72 FLVCR1feline leukemia virus subgroup C cellular receptor 1 (ENSG00000162769), score: 0.53 FNDC7fibronectin type III domain containing 7 (ENSG00000143107), score: 0.59 FUBP1far upstream element (FUSE) binding protein 1 (ENSG00000162613), score: 0.63 FUT9fucosyltransferase 9 (alpha (1,3) fucosyltransferase) (ENSG00000172461), score: 0.5 GAAglucosidase, alpha; acid (ENSG00000171298), score: -0.62 GABRB2gamma-aminobutyric acid (GABA) A receptor, beta 2 (ENSG00000145864), score: 0.46 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.48 GAD2glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) (ENSG00000136750), score: 0.5 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (ENSG00000141429), score: 0.52 GDF10growth differentiation factor 10 (ENSG00000107623), score: 0.53 GFM2G elongation factor, mitochondrial 2 (ENSG00000164347), score: -0.46 GHITMgrowth hormone inducible transmembrane protein (ENSG00000165678), score: -0.49 GLT8D1glycosyltransferase 8 domain containing 1 (ENSG00000016864), score: -0.48 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: -0.45 GNAI1guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 (ENSG00000127955), score: 0.45 GNG13guanine nucleotide binding protein (G protein), gamma 13 (ENSG00000127588), score: 0.63 GNPTGN-acetylglucosamine-1-phosphate transferase, gamma subunit (ENSG00000090581), score: -0.44 GPR149G protein-coupled receptor 149 (ENSG00000174948), score: 0.59 GPR37L1G protein-coupled receptor 37 like 1 (ENSG00000170075), score: 0.5 GPR85G protein-coupled receptor 85 (ENSG00000164604), score: 0.53 GRAMD2GRAM domain containing 2 (ENSG00000175318), score: 0.5 GRAMD3GRAM domain containing 3 (ENSG00000155324), score: -0.52 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.76 GRIA4glutamate receptor, ionotrophic, AMPA 4 (ENSG00000152578), score: 0.47 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.86 GRIK1glutamate receptor, ionotropic, kainate 1 (ENSG00000171189), score: 0.66 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.68 H2AFY2H2A histone family, member Y2 (ENSG00000099284), score: 0.5 HCN1hyperpolarization activated cyclic nucleotide-gated potassium channel 1 (ENSG00000164588), score: 0.45 HDXhighly divergent homeobox (ENSG00000165259), score: 0.52 HEATR5BHEAT repeat containing 5B (ENSG00000008869), score: 0.47 IBSPintegrin-binding sialoprotein (ENSG00000029559), score: 0.55 IFFO1intermediate filament family orphan 1 (ENSG00000010295), score: 0.47 IL10RBinterleukin 10 receptor, beta (ENSG00000243646), score: -0.47 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.59 INPPL1inositol polyphosphate phosphatase-like 1 (ENSG00000165458), score: -0.46 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: 0.51 JARID2jumonji, AT rich interactive domain 2 (ENSG00000008083), score: 0.5 KBTBD3kelch repeat and BTB (POZ) domain containing 3 (ENSG00000182359), score: 0.52 KCNA1potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) (ENSG00000111262), score: 0.47 KCNA10potassium voltage-gated channel, shaker-related subfamily, member 10 (ENSG00000143105), score: 0.78 KCNC1potassium voltage-gated channel, Shaw-related subfamily, member 1 (ENSG00000129159), score: 0.5 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (ENSG00000171385), score: 0.45 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.68 KERAkeratocan (ENSG00000139330), score: 0.78 KIAA0174KIAA0174 (ENSG00000182149), score: -0.45 KIAA0391KIAA0391 (ENSG00000100890), score: -0.44 KIAA2022KIAA2022 (ENSG00000050030), score: 0.57 KLF6Kruppel-like factor 6 (ENSG00000067082), score: -0.5 KLHDC3kelch domain containing 3 (ENSG00000124702), score: -0.46 LAMB1laminin, beta 1 (ENSG00000091136), score: -0.45 LANCL2LanC lantibiotic synthetase component C-like 2 (bacterial) (ENSG00000132434), score: 0.5 LARP4BLa ribonucleoprotein domain family, member 4B (ENSG00000107929), score: -0.61 LEMD2LEM domain containing 2 (ENSG00000161904), score: -0.5 LFNGLFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (ENSG00000106003), score: 0.46 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.74 LIN7Alin-7 homolog A (C. elegans) (ENSG00000111052), score: 0.52 LOC100132308solute carrier family 29 (nucleoside transporters), member 4 pseudogene (ENSG00000164638), score: 0.52 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.59 LOC100293817similar to hCG1811893 (ENSG00000144460), score: 0.63 LONP1lon peptidase 1, mitochondrial (ENSG00000196365), score: -0.5 LONRF3LON peptidase N-terminal domain and ring finger 3 (ENSG00000175556), score: 0.58 LPHN3latrophilin 3 (ENSG00000150471), score: 0.46 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (ENSG00000126243), score: 0.58 LRP12low density lipoprotein receptor-related protein 12 (ENSG00000147650), score: 0.45 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.56 LRRC33leucine rich repeat containing 33 (ENSG00000174004), score: -0.54 LRRC38leucine rich repeat containing 38 (ENSG00000162494), score: 0.67 LRRN1leucine rich repeat neuronal 1 (ENSG00000175928), score: 0.57 LSAMPlimbic system-associated membrane protein (ENSG00000185565), score: 0.48 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: -0.49 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.76 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.58 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.83 MAMLD1mastermind-like domain containing 1 (ENSG00000013619), score: 0.63 MAN2A1mannosidase, alpha, class 2A, member 1 (ENSG00000112893), score: -0.46 MAP3K13mitogen-activated protein kinase kinase kinase 13 (ENSG00000073803), score: 0.48 MAP3K15mitogen-activated protein kinase kinase kinase 15 (ENSG00000180815), score: -0.45 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: -0.48 MARCH6membrane-associated ring finger (C3HC4) 6 (ENSG00000145495), score: -0.45 MCTP1multiple C2 domains, transmembrane 1 (ENSG00000175471), score: 0.48 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.56 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.62 MIIPmigration and invasion inhibitory protein (ENSG00000116691), score: -0.5 MIXL1Mix1 homeobox-like 1 (Xenopus laevis) (ENSG00000185155), score: 0.82 MLH1mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) (ENSG00000076242), score: 0.46 MMP16matrix metallopeptidase 16 (membrane-inserted) (ENSG00000156103), score: 0.59 MRPL28mitochondrial ribosomal protein L28 (ENSG00000086504), score: -0.5 MRPL51mitochondrial ribosomal protein L51 (ENSG00000111639), score: -0.51 MRPS9mitochondrial ribosomal protein S9 (ENSG00000135972), score: -0.44 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: -0.47 MTMR3myotubularin related protein 3 (ENSG00000100330), score: 0.6 MYO16myosin XVI (ENSG00000041515), score: 0.58 MYO5Cmyosin VC (ENSG00000128833), score: -0.46 MYO9Bmyosin IXB (ENSG00000099331), score: -0.45 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.48 NARFLnuclear prelamin A recognition factor-like (ENSG00000103245), score: -0.46 NECAP2NECAP endocytosis associated 2 (ENSG00000157191), score: -0.47 NFIAnuclear factor I/A (ENSG00000162599), score: 0.45 NFRKBnuclear factor related to kappaB binding protein (ENSG00000170322), score: 0.51 NGBneuroglobin (ENSG00000165553), score: 0.71 NKAIN4Na+/K+ transporting ATPase interacting 4 (ENSG00000101198), score: 0.5 NMNAT2nicotinamide nucleotide adenylyltransferase 2 (ENSG00000157064), score: 0.48 NRG2neuregulin 2 (ENSG00000158458), score: 0.58 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: -0.46 NTMneurotrimin (ENSG00000182667), score: 0.48 NTRK3neurotrophic tyrosine kinase, receptor, type 3 (ENSG00000140538), score: 0.51 OCIAD1OCIA domain containing 1 (ENSG00000109180), score: -0.6 PAFAH1B2platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) (ENSG00000168092), score: 0.46 PAK7p21 protein (Cdc42/Rac)-activated kinase 7 (ENSG00000101349), score: 0.57 PAX6paired box 6 (ENSG00000007372), score: 0.63 PCGF3polycomb group ring finger 3 (ENSG00000185619), score: -0.53 PCID2PCI domain containing 2 (ENSG00000126226), score: -0.51 PCSK2proprotein convertase subtilisin/kexin type 2 (ENSG00000125851), score: 0.46 PDCLphosducin-like (ENSG00000136940), score: 0.53 PDE5Aphosphodiesterase 5A, cGMP-specific (ENSG00000138735), score: 0.53 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.45 PHF16PHD finger protein 16 (ENSG00000102221), score: 0.51 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (ENSG00000171608), score: 0.56 PIK3IP1phosphoinositide-3-kinase interacting protein 1 (ENSG00000100100), score: 0.46 PIK3R3phosphoinositide-3-kinase, regulatory subunit 3 (gamma) (ENSG00000117461), score: 0.5 PLCB4phospholipase C, beta 4 (ENSG00000101333), score: 0.54 PLCXD3phosphatidylinositol-specific phospholipase C, X domain containing 3 (ENSG00000182836), score: 0.5 PLD5phospholipase D family, member 5 (ENSG00000180287), score: 0.46 PPP3CAprotein phosphatase 3, catalytic subunit, alpha isozyme (ENSG00000138814), score: 0.48 PPP4R4protein phosphatase 4, regulatory subunit 4 (ENSG00000119698), score: 0.47 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.58 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.46 PRDM11PR domain containing 11 (ENSG00000019485), score: 0.52 PSD2pleckstrin and Sec7 domain containing 2 (ENSG00000146005), score: 0.51 PSEN1presenilin 1 (ENSG00000080815), score: -0.48 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.54 PTP4A2protein tyrosine phosphatase type IVA, member 2 (ENSG00000184007), score: -0.7 PTPREprotein tyrosine phosphatase, receptor type, E (ENSG00000132334), score: 0.48 PTPRRprotein tyrosine phosphatase, receptor type, R (ENSG00000153233), score: 0.56 PVALBparvalbumin (ENSG00000100362), score: 0.56 RAB30RAB30, member RAS oncogene family (ENSG00000137502), score: 0.57 RAB32RAB32, member RAS oncogene family (ENSG00000118508), score: -0.45 RAB3GAP2RAB3 GTPase activating protein subunit 2 (non-catalytic) (ENSG00000118873), score: 0.49 RAB9BRAB9B, member RAS oncogene family (ENSG00000123570), score: 0.47 RBM45RNA binding motif protein 45 (ENSG00000155636), score: 0.48 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.53 RGS2regulator of G-protein signaling 2, 24kDa (ENSG00000116741), score: -0.48 RGS7BPregulator of G-protein signaling 7 binding protein (ENSG00000186479), score: 0.51 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.57 RNF114ring finger protein 114 (ENSG00000124226), score: -0.52 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.49 RXRAretinoid X receptor, alpha (ENSG00000186350), score: -0.62 SAMD12sterile alpha motif domain containing 12 (ENSG00000177570), score: 0.5 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: 0.48 SEPT6septin 6 (ENSG00000125354), score: 0.46 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (ENSG00000197632), score: 0.74 SESTD1SEC14 and spectrin domains 1 (ENSG00000187231), score: 0.53 SGCZsarcoglycan, zeta (ENSG00000185053), score: 0.66 SGSM1small G protein signaling modulator 1 (ENSG00000167037), score: 0.52 SGTBsmall glutamine-rich tetratricopeptide repeat (TPR)-containing, beta (ENSG00000197860), score: 0.46 SH2B2SH2B adaptor protein 2 (ENSG00000160999), score: 0.45 SHOC2soc-2 suppressor of clear homolog (C. elegans) (ENSG00000108061), score: -0.5 SKOR1SKI family transcriptional corepressor 1 (ENSG00000188779), score: 0.69 SLC12A5solute carrier family 12 (potassium/chloride transporter), member 5 (ENSG00000124140), score: 0.45 SLC24A5solute carrier family 24, member 5 (ENSG00000188467), score: 0.5 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.57 SLC35A5solute carrier family 35, member A5 (ENSG00000138459), score: -0.45 SLC35B3solute carrier family 35, member B3 (ENSG00000124786), score: -0.48 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.65 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.63 SNAP23synaptosomal-associated protein, 23kDa (ENSG00000092531), score: -0.52 SNRKSNF related kinase (ENSG00000163788), score: 0.67 SNX25sorting nexin 25 (ENSG00000109762), score: 0.73 SNX3sorting nexin 3 (ENSG00000112335), score: -0.6 SNX8sorting nexin 8 (ENSG00000106266), score: -0.51 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (ENSG00000090487), score: -0.58 SPHKAPSPHK1 interactor, AKAP domain containing (ENSG00000153820), score: 0.55 SPON1spondin 1, extracellular matrix protein (ENSG00000152268), score: 0.57 SRRM4serine/arginine repetitive matrix 4 (ENSG00000139767), score: 0.46 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.57 STX12syntaxin 12 (ENSG00000117758), score: 0.54 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.46 SUSD4sushi domain containing 4 (ENSG00000143502), score: 0.45 SYBUsyntabulin (syntaxin-interacting) (ENSG00000147642), score: 0.5 SYNJ1synaptojanin 1 (ENSG00000159082), score: 0.45 TT, brachyury homolog (mouse) (ENSG00000164458), score: 0.53 TAPT1transmembrane anterior posterior transformation 1 (ENSG00000169762), score: -0.45 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: 0.52 TFAP2Atranscription factor AP-2 alpha (activating enhancer binding protein 2 alpha) (ENSG00000137203), score: 0.46 TFAP2Btranscription factor AP-2 beta (activating enhancer binding protein 2 beta) (ENSG00000008196), score: 0.49 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (ENSG00000198959), score: -0.53 THSD7Athrombospondin, type I, domain containing 7A (ENSG00000005108), score: 0.53 TIMM44translocase of inner mitochondrial membrane 44 homolog (yeast) (ENSG00000104980), score: -0.52 TLL1tolloid-like 1 (ENSG00000038295), score: 0.77 TMEM120Btransmembrane protein 120B (ENSG00000188735), score: 0.54 TMEM22transmembrane protein 22 (ENSG00000168917), score: 0.57 TMEM43transmembrane protein 43 (ENSG00000170876), score: -0.49 TMEM63Ctransmembrane protein 63C (ENSG00000165548), score: 0.51 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (ENSG00000185215), score: -0.48 TNKStankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase (ENSG00000173273), score: 0.51 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: -0.52 TRAF5TNF receptor-associated factor 5 (ENSG00000082512), score: -0.44 TRDMT1tRNA aspartic acid methyltransferase 1 (ENSG00000107614), score: 0.45 TRIM3tripartite motif-containing 3 (ENSG00000110171), score: 0.49 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.78 TRPC1transient receptor potential cation channel, subfamily C, member 1 (ENSG00000144935), score: 0.49 TRPC3transient receptor potential cation channel, subfamily C, member 3 (ENSG00000138741), score: 0.86 TUBGCP4tubulin, gamma complex associated protein 4 (ENSG00000137822), score: 0.48 TXNL1thioredoxin-like 1 (ENSG00000091164), score: -0.45 UPF0639UPF0639 protein (ENSG00000175985), score: 0.82 USP20ubiquitin specific peptidase 20 (ENSG00000136878), score: 0.46 WDFY3WD repeat and FYVE domain containing 3 (ENSG00000163625), score: 0.59 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.52 WNT16wingless-type MMTV integration site family, member 16 (ENSG00000002745), score: 0.56 XPO5exportin 5 (ENSG00000124571), score: 0.47 ZC4H2zinc finger, C4H2 domain containing (ENSG00000126970), score: 0.49 ZDHHC15zinc finger, DHHC-type containing 15 (ENSG00000102383), score: 0.47 ZFPM2zinc finger protein, multitype 2 (ENSG00000169946), score: 0.57 ZIC4Zic family member 4 (ENSG00000174963), score: 0.82 ZNF238zinc finger protein 238 (ENSG00000179456), score: 0.6 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.45 ZNF385Bzinc finger protein 385B (ENSG00000144331), score: 0.46 ZNF385Czinc finger protein 385C (ENSG00000187595), score: 0.69 ZNF423zinc finger protein 423 (ENSG00000102935), score: 0.63 ZNF462zinc finger protein 462 (ENSG00000148143), score: 0.57 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.55 ZNF536zinc finger protein 536 (ENSG00000198597), score: 0.46
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_cb_m_ca1 | gga | cb | m | _ |
| gga_cb_f_ca1 | gga | cb | f | _ |
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |