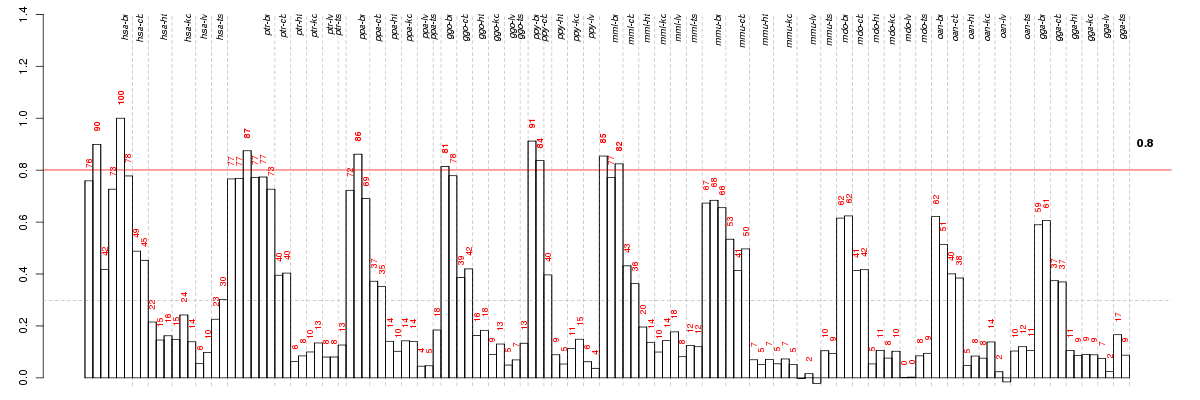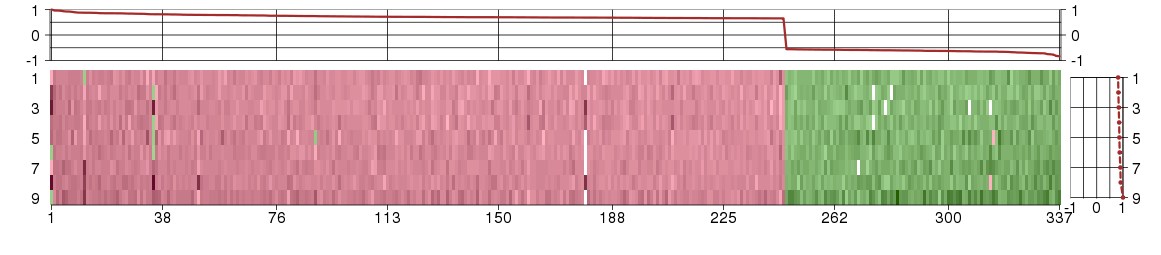

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cation transport
The directed movement of cations, atoms or small molecules with a net positive charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
potassium ion transport
The directed movement of potassium ions (K+) into, out of, within or between cells by means of some external agent such as a transporter or pore.
metal ion transport
The directed movement of metal ions, any metal ion with an electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
homophilic cell adhesion
The attachment of an adhesion molecule in one cell to an identical molecule in an adjacent cell.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
monovalent inorganic cation transport
The directed movement of inorganic cations with a valency of one into, out of, within or between cells by means of some external agent such as a transporter or pore. Inorganic cations are atoms or small molecules with a positive charge which do not contain carbon in covalent linkage.
cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
NA
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
potassium ion transport
The directed movement of potassium ions (K+) into, out of, within or between cells by means of some external agent such as a transporter or pore.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
postsynaptic density
The post synaptic density is a region that lies adjacent to the cytoplasmic face of the postsynaptic membrane at excitatory synapse. It forms a disc that consists of a range of proteins with different functions, some of which contact the cytoplasmic domains of ion channels in the postsynaptic membrane. The proteins making up the disc include receptors, and structural proteins linked to the actin cytoskeleton. They also include signalling machinery, such as protein kinases and phosphatases.
cell junction
A plasma membrane part that forms a specialized region of connection between two cells or between a cell and the extracellular matrix. At a cell junction, anchoring proteins extend through the plasma membrane to link cytoskeletal proteins in one cell to cytoskeletal proteins in neighboring cells or to proteins in the extracellular matrix.
dendrite
A neuron projection that has a short, tapering, often branched, morphology, receives and integrates signals from other neurons or from sensory stimuli, and conducts a nerve impulse towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
potassium channel complex
An ion channel complex through which potassium ions pass.
presynaptic membrane
A specialized area of membrane of the axon terminal that faces the plasma membrane of the neuron or muscle fiber with which the axon terminal establishes a synaptic junction; many synaptic junctions exhibit structural presynaptic characteristics, such as conical, electron-dense internal protrusions, that distinguish it from the remainder of the axon plasma membrane.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
neuron projection
A prolongation or process extending from a nerve cell, e.g. an axon or dendrite.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse
The junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell; the site of interneuronal communication. As the nerve fiber approaches the synapse it enlarges into a specialized structure, the presynaptic nerve ending, which contains mitochondria and synaptic vesicles. At the tip of the nerve ending is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic nerve ending secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane.
postsynaptic membrane
A specialized area of membrane facing the presynaptic membrane on the tip of the nerve ending and separated from it by a minute cleft (the synaptic cleft). Neurotransmitters across the synaptic cleft and transmit the signal to the postsynaptic membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
presynaptic membrane
A specialized area of membrane of the axon terminal that faces the plasma membrane of the neuron or muscle fiber with which the axon terminal establishes a synaptic junction; many synaptic junctions exhibit structural presynaptic characteristics, such as conical, electron-dense internal protrusions, that distinguish it from the remainder of the axon plasma membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
postsynaptic membrane
A specialized area of membrane facing the presynaptic membrane on the tip of the nerve ending and separated from it by a minute cleft (the synaptic cleft). Neurotransmitters across the synaptic cleft and transmit the signal to the postsynaptic membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
postsynaptic density
The post synaptic density is a region that lies adjacent to the cytoplasmic face of the postsynaptic membrane at excitatory synapse. It forms a disc that consists of a range of proteins with different functions, some of which contact the cytoplasmic domains of ion channels in the postsynaptic membrane. The proteins making up the disc include receptors, and structural proteins linked to the actin cytoskeleton. They also include signalling machinery, such as protein kinases and phosphatases.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
protein kinase activity
Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP.
protein serine/threonine kinase activity
Catalysis of the reaction: ATP + a protein serine/threonine = ADP + protein serine/threonine phosphate.
calmodulin-dependent protein kinase activity
Catalysis of the reaction: ATP + a protein serine/threonine = ADP + protein serine/threonine phosphate, dependent on the presence of calcium-bound calmodulin.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
voltage-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons.
voltage-gated potassium channel activity
Catalysis of the transmembrane transfer of a potassium ion by a voltage-gated channel.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
potassium channel activity
Catalysis of facilitated diffusion of a potassium ion (by an energy-independent process) involving passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
glutamate receptor activity
Combining with glutamate to initiate a change in cell activity.
G-protein coupled amine receptor activity
A receptor that binds an extracellular amine and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
kinase activity
Catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
transferase activity
Catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
transferase activity, transferring phosphorus-containing groups
Catalysis of the transfer of a phosphorus-containing group from one compound (donor) to another (acceptor).
phosphotransferase activity, alcohol group as acceptor
Catalysis of the transfer of a phosphorus-containing group from one compound (donor) to an alcohol group (acceptor).
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
voltage-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel whose open state is dependent on the voltage across the membrane in which it is embedded.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
voltage-gated cation channel activity
Catalysis of the transmembrane transfer of a cation by a voltage-gated channel. A cation is a positively charged ion.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
protein kinase activity
Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
voltage-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons.
voltage-gated cation channel activity
Catalysis of the transmembrane transfer of a cation by a voltage-gated channel. A cation is a positively charged ion.
voltage-gated potassium channel activity
Catalysis of the transmembrane transfer of a potassium ion by a voltage-gated channel.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 4.262e-03 | 6.09 | 17 | 102 | Neuroactive ligand-receptor interaction |
| 04020 | 2.151e-02 | 4.06 | 12 | 68 | Calcium signaling pathway |
ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: -0.62 ABHD12abhydrolase domain containing 12 (ENSG00000100997), score: 0.69 ABRactive BCR-related gene (ENSG00000159842), score: 0.74 ACTL6Aactin-like 6A (ENSG00000136518), score: -0.7 ACTN1actinin, alpha 1 (ENSG00000072110), score: 0.67 ADAMTS8ADAM metallopeptidase with thrombospondin type 1 motif, 8 (ENSG00000134917), score: 0.78 ADCY2adenylate cyclase 2 (brain) (ENSG00000078295), score: 0.72 AIMP1aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 (ENSG00000164022), score: -0.58 AMZ1archaelysin family metallopeptidase 1 (ENSG00000174945), score: 0.65 ANKRD24ankyrin repeat domain 24 (ENSG00000089847), score: 0.75 ANKRD33Bankyrin repeat domain 33B (ENSG00000164236), score: 0.66 AP3B1adaptor-related protein complex 3, beta 1 subunit (ENSG00000132842), score: -0.61 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: -0.57 ARCactivity-regulated cytoskeleton-associated protein (ENSG00000198576), score: 0.78 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: -0.72 ARHGAP32Rho GTPase activating protein 32 (ENSG00000134909), score: 0.66 ARNTaryl hydrocarbon receptor nuclear translocator (ENSG00000143437), score: -0.58 ASB6ankyrin repeat and SOCS box-containing 6 (ENSG00000148331), score: 0.79 ATG4BATG4 autophagy related 4 homolog B (S. cerevisiae) (ENSG00000168397), score: 0.66 ATP13A2ATPase type 13A2 (ENSG00000159363), score: 0.78 BAI1brain-specific angiogenesis inhibitor 1 (ENSG00000181790), score: 0.79 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (ENSG00000163930), score: 0.86 BCL11BB-cell CLL/lymphoma 11B (zinc finger protein) (ENSG00000127152), score: 0.67 BEGAINbrain-enriched guanylate kinase-associated homolog (rat) (ENSG00000183092), score: 0.72 BTBD2BTB (POZ) domain containing 2 (ENSG00000133243), score: 0.69 C11orf41chromosome 11 open reading frame 41 (ENSG00000110427), score: 0.66 C12orf26chromosome 12 open reading frame 26 (ENSG00000127720), score: -0.57 C12orf53chromosome 12 open reading frame 53 (ENSG00000139200), score: 0.7 C13orf36chromosome 13 open reading frame 36 (ENSG00000180440), score: 0.83 C1orf128chromosome 1 open reading frame 128 (ENSG00000057757), score: 0.7 C1orf55chromosome 1 open reading frame 55 (ENSG00000143751), score: -0.62 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.72 C6orf168chromosome 6 open reading frame 168 (ENSG00000146267), score: 0.69 C8orf46chromosome 8 open reading frame 46 (ENSG00000169085), score: 0.81 C9orf16chromosome 9 open reading frame 16 (ENSG00000171159), score: 0.77 CA7carbonic anhydrase VII (ENSG00000168748), score: 0.7 CABP1calcium binding protein 1 (ENSG00000157782), score: 0.68 CACNA1Bcalcium channel, voltage-dependent, N type, alpha 1B subunit (ENSG00000148408), score: 0.65 CACNA1Ecalcium channel, voltage-dependent, R type, alpha 1E subunit (ENSG00000198216), score: 0.68 CACNG3calcium channel, voltage-dependent, gamma subunit 3 (ENSG00000006116), score: 0.75 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 0.87 CAMK1Dcalcium/calmodulin-dependent protein kinase ID (ENSG00000183049), score: 0.66 CAMK1Gcalcium/calmodulin-dependent protein kinase IG (ENSG00000008118), score: 0.79 CAMK2Acalcium/calmodulin-dependent protein kinase II alpha (ENSG00000070808), score: 0.79 CAMK2Gcalcium/calmodulin-dependent protein kinase II gamma (ENSG00000148660), score: 0.69 CBLN4cerebellin 4 precursor (ENSG00000054803), score: 0.92 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: -0.62 CCDC3coiled-coil domain containing 3 (ENSG00000151468), score: 0.67 CCDC50coiled-coil domain containing 50 (ENSG00000152492), score: -0.61 CCDC85Acoiled-coil domain containing 85A (ENSG00000055813), score: 0.7 CCM2cerebral cavernous malformation 2 (ENSG00000136280), score: 0.68 CD2APCD2-associated protein (ENSG00000198087), score: -0.62 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: 0.65 CDH10cadherin 10, type 2 (T2-cadherin) (ENSG00000040731), score: 0.65 CDH12cadherin 12, type 2 (N-cadherin 2) (ENSG00000154162), score: 0.72 CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.75 CDH9cadherin 9, type 2 (T1-cadherin) (ENSG00000113100), score: 0.69 CELF5CUGBP, Elav-like family member 5 (ENSG00000161082), score: 0.72 CETN3centrin, EF-hand protein, 3 (ENSG00000153140), score: -0.64 CHN1chimerin (chimaerin) 1 (ENSG00000128656), score: 0.7 CHRM3cholinergic receptor, muscarinic 3 (ENSG00000133019), score: 0.85 CHRNA4cholinergic receptor, nicotinic, alpha 4 (ENSG00000101204), score: 0.65 CHST1carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 (ENSG00000175264), score: 0.78 CLN5ceroid-lipofuscinosis, neuronal 5 (ENSG00000102805), score: -0.57 CNIH3cornichon homolog 3 (Drosophila) (ENSG00000143786), score: 0.7 COPB1coatomer protein complex, subunit beta 1 (ENSG00000129083), score: -0.68 CPNE4copine IV (ENSG00000196353), score: 0.68 CRHcorticotropin releasing hormone (ENSG00000147571), score: 0.68 CRLS1cardiolipin synthase 1 (ENSG00000088766), score: -0.57 CRTAC1cartilage acidic protein 1 (ENSG00000095713), score: 0.66 CTNND2catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein) (ENSG00000169862), score: 0.71 DCLK1doublecortin-like kinase 1 (ENSG00000133083), score: 0.68 DERL2Der1-like domain family, member 2 (ENSG00000072849), score: -0.59 DGCR2DiGeorge syndrome critical region gene 2 (ENSG00000070413), score: 0.81 DGKZdiacylglycerol kinase, zeta 104kDa (ENSG00000149091), score: 0.83 DLGAP2discs, large (Drosophila) homolog-associated protein 2 (ENSG00000198010), score: 0.77 DLX1distal-less homeobox 1 (ENSG00000144355), score: 0.89 DNAH5dynein, axonemal, heavy chain 5 (ENSG00000039139), score: 0.68 DRP2dystrophin related protein 2 (ENSG00000102385), score: 0.7 DSCAMDown syndrome cell adhesion molecule (ENSG00000171587), score: 0.73 DSCAML1Down syndrome cell adhesion molecule like 1 (ENSG00000177103), score: 0.68 EIF3Eeukaryotic translation initiation factor 3, subunit E (ENSG00000104408), score: -0.58 EIF3Meukaryotic translation initiation factor 3, subunit M (ENSG00000149100), score: -0.56 ELFN2extracellular leucine-rich repeat and fibronectin type III domain containing 2 (ENSG00000166897), score: 0.8 ELMOD2ELMO/CED-12 domain containing 2 (ENSG00000179387), score: -0.82 ENC1ectodermal-neural cortex 1 (with BTB-like domain) (ENSG00000171617), score: 0.87 ENTPD3ectonucleoside triphosphate diphosphohydrolase 3 (ENSG00000168032), score: 0.69 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: -0.6 EPHB6EPH receptor B6 (ENSG00000106123), score: 0.87 ERI1exoribonuclease 1 (ENSG00000104626), score: -0.6 ETAA1Ewing tumor-associated antigen 1 (ENSG00000143971), score: -0.7 EXOC7exocyst complex component 7 (ENSG00000182473), score: 0.71 FAM102Afamily with sequence similarity 102, member A (ENSG00000167106), score: 0.68 FAM168Bfamily with sequence similarity 168, member B (ENSG00000152102), score: 0.71 FAM189A1family with sequence similarity 189, member A1 (ENSG00000104059), score: 0.67 FAM19A1family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 (ENSG00000183662), score: 0.87 FAM49Afamily with sequence similarity 49, member A (ENSG00000197872), score: 0.66 FAM5Bfamily with sequence similarity 5, member B (ENSG00000198797), score: 0.79 FAM60Afamily with sequence similarity 60, member A (ENSG00000139146), score: -0.63 FAM81Afamily with sequence similarity 81, member A (ENSG00000157470), score: 0.68 FBLN7fibulin 7 (ENSG00000144152), score: 0.84 FBXL16F-box and leucine-rich repeat protein 16 (ENSG00000127585), score: 0.75 FBXL5F-box and leucine-rich repeat protein 5 (ENSG00000118564), score: -0.58 FBXO8F-box protein 8 (ENSG00000164117), score: -0.66 FCHO2FCH domain only 2 (ENSG00000157107), score: -0.64 FERD3LFer3-like (Drosophila) (ENSG00000146618), score: 1 FEZF2FEZ family zinc finger 2 (ENSG00000153266), score: 0.91 FLRT2fibronectin leucine rich transmembrane protein 2 (ENSG00000185070), score: 0.7 FN3KRPfructosamine 3 kinase related protein (ENSG00000141560), score: 0.66 FOXG1forkhead box G1 (ENSG00000176165), score: 0.88 GABPAGA binding protein transcription factor, alpha subunit 60kDa (ENSG00000154727), score: -0.66 GABRB3gamma-aminobutyric acid (GABA) A receptor, beta 3 (ENSG00000166206), score: 0.7 GLRA3glycine receptor, alpha 3 (ENSG00000145451), score: 0.69 GNAZguanine nucleotide binding protein (G protein), alpha z polypeptide (ENSG00000128266), score: 0.68 GNB1guanine nucleotide binding protein (G protein), beta polypeptide 1 (ENSG00000078369), score: 0.75 GOLGA4golgin A4 (ENSG00000144674), score: -0.75 GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.93 GPR176G protein-coupled receptor 176 (ENSG00000166073), score: 0.77 GPR83G protein-coupled receptor 83 (ENSG00000123901), score: 0.75 GRIA3glutamate receptor, ionotrophic, AMPA 3 (ENSG00000125675), score: 0.69 GRID1glutamate receptor, ionotropic, delta 1 (ENSG00000182771), score: 0.69 GRIK4glutamate receptor, ionotropic, kainate 4 (ENSG00000149403), score: 0.67 GRIN1glutamate receptor, ionotropic, N-methyl D-aspartate 1 (ENSG00000176884), score: 0.71 GRIN2Aglutamate receptor, ionotropic, N-methyl D-aspartate 2A (ENSG00000183454), score: 0.74 GRM3glutamate receptor, metabotropic 3 (ENSG00000198822), score: 0.66 GTF2A2general transcription factor IIA, 2, 12kDa (ENSG00000140307), score: -0.56 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: -0.64 HAT1histone acetyltransferase 1 (ENSG00000128708), score: -0.71 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: -0.62 HCN1hyperpolarization activated cyclic nucleotide-gated potassium channel 1 (ENSG00000164588), score: 0.68 HEATR5AHEAT repeat containing 5A (ENSG00000129493), score: -0.59 HIBCH3-hydroxyisobutyryl-CoA hydrolase (ENSG00000198130), score: -0.57 HMGXB3HMG box domain containing 3 (ENSG00000113716), score: 0.68 HRH2histamine receptor H2 (ENSG00000113749), score: 0.7 HRH3histamine receptor H3 (ENSG00000101180), score: 0.77 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (ENSG00000102468), score: 0.95 HTR45-hydroxytryptamine (serotonin) receptor 4 (ENSG00000164270), score: 0.69 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (ENSG00000005700), score: -0.58 IDEinsulin-degrading enzyme (ENSG00000119912), score: -0.65 IGSF21immunoglobin superfamily, member 21 (ENSG00000117154), score: 0.66 INTS8integrator complex subunit 8 (ENSG00000164941), score: -0.58 ITPK1inositol 1,3,4-triphosphate 5/6 kinase (ENSG00000100605), score: 0.74 ITPKAinositol 1,4,5-trisphosphate 3-kinase A (ENSG00000137825), score: 0.73 KCNA4potassium voltage-gated channel, shaker-related subfamily, member 4 (ENSG00000182255), score: 0.66 KCNA6potassium voltage-gated channel, shaker-related subfamily, member 6 (ENSG00000151079), score: 0.77 KCNF1potassium voltage-gated channel, subfamily F, member 1 (ENSG00000162975), score: 0.96 KCNG1potassium voltage-gated channel, subfamily G, member 1 (ENSG00000026559), score: 0.85 KCNG3potassium voltage-gated channel, subfamily G, member 3 (ENSG00000171126), score: 0.82 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (ENSG00000135643), score: 0.73 KCNQ2potassium voltage-gated channel, KQT-like subfamily, member 2 (ENSG00000075043), score: 0.72 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (ENSG00000184156), score: 0.71 KCNQ5potassium voltage-gated channel, KQT-like subfamily, member 5 (ENSG00000185760), score: 0.87 KCNS1potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 (ENSG00000124134), score: 0.96 KCNS2potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 (ENSG00000156486), score: 0.83 KCTD1potassium channel tetramerisation domain containing 1 (ENSG00000134504), score: 0.66 KCTD16potassium channel tetramerisation domain containing 16 (ENSG00000183775), score: 0.67 KCTD4potassium channel tetramerisation domain containing 4 (ENSG00000180332), score: 0.7 KDM5Alysine (K)-specific demethylase 5A (ENSG00000073614), score: -0.63 KIAA0020KIAA0020 (ENSG00000080608), score: -0.66 KIAA0284KIAA0284 (ENSG00000099814), score: 0.72 KIAA0427KIAA0427 (ENSG00000134030), score: 0.67 KIAA1033KIAA1033 (ENSG00000136051), score: -0.59 KIF21Bkinesin family member 21B (ENSG00000116852), score: 0.69 KIRREL3kin of IRRE like 3 (Drosophila) (ENSG00000149571), score: 0.71 KLHDC3kelch domain containing 3 (ENSG00000124702), score: 0.67 KLHL2kelch-like 2, Mayven (Drosophila) (ENSG00000109466), score: 0.68 KLHL22kelch-like 22 (Drosophila) (ENSG00000099910), score: 0.7 LACTBlactamase, beta (ENSG00000103642), score: -0.56 LAPTM4Alysosomal protein transmembrane 4 alpha (ENSG00000068697), score: -0.65 LBRlamin B receptor (ENSG00000143815), score: -0.63 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: -0.64 LIMA1LIM domain and actin binding 1 (ENSG00000050405), score: -0.58 LIMK1LIM domain kinase 1 (ENSG00000106683), score: 0.75 LINGO2leucine rich repeat and Ig domain containing 2 (ENSG00000174482), score: 0.72 LMO4LIM domain only 4 (ENSG00000143013), score: 0.88 LMTK2lemur tyrosine kinase 2 (ENSG00000164715), score: 0.81 LOC100132308solute carrier family 29 (nucleoside transporters), member 4 pseudogene (ENSG00000164638), score: 0.71 LPCAT3lysophosphatidylcholine acyltransferase 3 (ENSG00000111684), score: -0.57 LPPR3lipid phosphate phosphatase-related protein type 3 (ENSG00000129951), score: 0.85 LPPR4lipid phosphate phosphatase-related protein type 4 (ENSG00000117600), score: 0.75 LRFN2leucine rich repeat and fibronectin type III domain containing 2 (ENSG00000156564), score: 0.92 LRP3low density lipoprotein receptor-related protein 3 (ENSG00000130881), score: 0.67 LRRC4Cleucine rich repeat containing 4C (ENSG00000148948), score: 0.67 LRTM2leucine-rich repeats and transmembrane domains 2 (ENSG00000166159), score: 0.8 LYSMD3LysM, putative peptidoglycan-binding, domain containing 3 (ENSG00000176018), score: -0.6 LZICleucine zipper and CTNNBIP1 domain containing (ENSG00000162441), score: -0.67 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (ENSG00000197063), score: 0.68 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: -0.69 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (ENSG00000138834), score: 0.69 MARCH7membrane-associated ring finger (C3HC4) 7 (ENSG00000136536), score: -0.6 MATKmegakaryocyte-associated tyrosine kinase (ENSG00000007264), score: 0.85 MATN1matrilin 1, cartilage matrix protein (ENSG00000162510), score: 0.69 MC4Rmelanocortin 4 receptor (ENSG00000166603), score: 0.79 MCEEmethylmalonyl CoA epimerase (ENSG00000124370), score: -0.56 MDM4Mdm4 p53 binding protein homolog (mouse) (ENSG00000198625), score: -0.65 MED7mediator complex subunit 7 (ENSG00000155868), score: -0.59 MEF2Cmyocyte enhancer factor 2C (ENSG00000081189), score: 0.74 MGAT5Bmannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B (ENSG00000167889), score: 0.81 MKL1megakaryoblastic leukemia (translocation) 1 (ENSG00000196588), score: 0.85 MKL2MKL/myocardin-like 2 (ENSG00000186260), score: 0.77 MRPL50mitochondrial ribosomal protein L50 (ENSG00000136897), score: -0.6 MTFR1mitochondrial fission regulator 1 (ENSG00000066855), score: -0.71 MYBPC1myosin binding protein C, slow type (ENSG00000196091), score: 0.67 MYNNmyoneurin (ENSG00000085274), score: -0.6 MYT1Lmyelin transcription factor 1-like (ENSG00000186487), score: 0.68 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (ENSG00000103174), score: 0.7 NCDNneurochondrin (ENSG00000020129), score: 0.76 NCS1neuronal calcium sensor 1 (ENSG00000107130), score: 0.71 NEDD1neural precursor cell expressed, developmentally down-regulated 1 (ENSG00000139350), score: -0.65 NELFnasal embryonic LHRH factor (ENSG00000165802), score: 0.71 NETO1neuropilin (NRP) and tolloid (TLL)-like 1 (ENSG00000166342), score: 0.84 NFU1NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) (ENSG00000169599), score: -0.63 NGEFneuronal guanine nucleotide exchange factor (ENSG00000066248), score: 0.73 NPTNneuroplastin (ENSG00000156642), score: 0.68 NUDCD3NudC domain containing 3 (ENSG00000015676), score: 0.71 NUP54nucleoporin 54kDa (ENSG00000138750), score: -0.56 OCA2oculocutaneous albinism II (ENSG00000104044), score: 0.85 OLFM1olfactomedin 1 (ENSG00000130558), score: 0.7 OPN4opsin 4 (ENSG00000122375), score: 0.81 OPRL1opiate receptor-like 1 (ENSG00000125510), score: 0.77 PALMparalemmin (ENSG00000099864), score: 0.69 PAN3PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (ENSG00000152520), score: -0.59 PANX2pannexin 2 (ENSG00000073150), score: 0.66 PCDH19protocadherin 19 (ENSG00000165194), score: 0.81 PCDH20protocadherin 20 (ENSG00000197991), score: 0.68 PCDH9protocadherin 9 (ENSG00000184226), score: 0.67 PCSK1proprotein convertase subtilisin/kexin type 1 (ENSG00000175426), score: 0.83 PDE8Bphosphodiesterase 8B (ENSG00000113231), score: 0.8 PDZD7PDZ domain containing 7 (ENSG00000186862), score: 0.74 PEX13peroxisomal biogenesis factor 13 (ENSG00000162928), score: -0.69 PHACTR4phosphatase and actin regulator 4 (ENSG00000204138), score: -0.62 PIK3C2Aphosphoinositide-3-kinase, class 2, alpha polypeptide (ENSG00000011405), score: -0.64 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (ENSG00000105647), score: 0.67 PKP1plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) (ENSG00000081277), score: 0.65 PLCH1phospholipase C, eta 1 (ENSG00000114805), score: 0.67 PLEKHM2pleckstrin homology domain containing, family M (with RUN domain) member 2 (ENSG00000116786), score: 0.72 PNOCprepronociceptin (ENSG00000168081), score: 0.68 POLR3Apolymerase (RNA) III (DNA directed) polypeptide A, 155kDa (ENSG00000148606), score: 0.78 PPARDperoxisome proliferator-activated receptor delta (ENSG00000112033), score: 0.7 PPFIA2protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 (ENSG00000139220), score: 0.67 PPME1protein phosphatase methylesterase 1 (ENSG00000214517), score: 0.65 PPP6R3protein phosphatase 6, regulatory subunit 3 (ENSG00000110075), score: -0.6 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.68 PRICKLE2prickle homolog 2 (Drosophila) (ENSG00000163637), score: 0.68 PRKAA1protein kinase, AMP-activated, alpha 1 catalytic subunit (ENSG00000132356), score: -0.61 PRKAR1Bprotein kinase, cAMP-dependent, regulatory, type I, beta (ENSG00000188191), score: 0.68 PRKAR2Bprotein kinase, cAMP-dependent, regulatory, type II, beta (ENSG00000005249), score: 0.69 PRPF19PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae) (ENSG00000110107), score: 0.74 PRPF40APRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) (ENSG00000196504), score: -0.61 PSDpleckstrin and Sec7 domain containing (ENSG00000059915), score: 0.77 PTHLHparathyroid hormone-like hormone (ENSG00000087494), score: 0.7 PTPRAprotein tyrosine phosphatase, receptor type, A (ENSG00000132670), score: 0.67 RAB11FIP4RAB11 family interacting protein 4 (class II) (ENSG00000131242), score: 0.66 RAB40CRAB40C, member RAS oncogene family (ENSG00000197562), score: 0.81 RAI1retinoic acid induced 1 (ENSG00000108557), score: 0.7 RANGAP1Ran GTPase activating protein 1 (ENSG00000100401), score: 0.67 RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.7 RASD1RAS, dexamethasone-induced 1 (ENSG00000108551), score: 0.66 RASD2RASD family, member 2 (ENSG00000100302), score: 0.68 RESTRE1-silencing transcription factor (ENSG00000084093), score: -0.59 REXO2REX2, RNA exonuclease 2 homolog (S. cerevisiae) (ENSG00000076043), score: -0.58 RHBDL3rhomboid, veinlet-like 3 (Drosophila) (ENSG00000141314), score: 0.71 RIMS3regulating synaptic membrane exocytosis 3 (ENSG00000117016), score: 0.78 RIMS4regulating synaptic membrane exocytosis 4 (ENSG00000101098), score: 0.66 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (ENSG00000067900), score: -0.83 RORBRAR-related orphan receptor B (ENSG00000198963), score: 0.81 RPRD1Aregulation of nuclear pre-mRNA domain containing 1A (ENSG00000141425), score: 0.69 RSPO2R-spondin 2 homolog (Xenopus laevis) (ENSG00000147655), score: 0.81 RTN4RL1reticulon 4 receptor-like 1 (ENSG00000185924), score: 0.73 SARSseryl-tRNA synthetase (ENSG00000031698), score: 0.7 SATB2SATB homeobox 2 (ENSG00000119042), score: 0.74 SCN3Bsodium channel, voltage-gated, type III, beta (ENSG00000166257), score: 0.73 SERPINI1serpin peptidase inhibitor, clade I (neuroserpin), member 1 (ENSG00000163536), score: 0.67 SGSM2small G protein signaling modulator 2 (ENSG00000141258), score: 0.71 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: 0.68 SIN3BSIN3 homolog B, transcription regulator (yeast) (ENSG00000127511), score: 0.78 SIRT1sirtuin 1 (ENSG00000096717), score: -0.58 SLC13A4solute carrier family 13 (sodium/sulfate symporters), member 4 (ENSG00000164707), score: 0.74 SLC24A4solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 (ENSG00000140090), score: 0.71 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (ENSG00000160326), score: 0.75 SLC6A7solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 (ENSG00000011083), score: 0.79 SMAD5SMAD family member 5 (ENSG00000113658), score: -0.59 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: -0.65 SNNstannin (ENSG00000184602), score: 0.7 SNTG1syntrophin, gamma 1 (ENSG00000147481), score: 0.71 SOS2son of sevenless homolog 2 (Drosophila) (ENSG00000100485), score: -0.59 SPDEFSAM pointed domain containing ets transcription factor (ENSG00000124664), score: 0.85 SREBF2sterol regulatory element binding transcription factor 2 (ENSG00000198911), score: 0.7 SSTsomatostatin (ENSG00000157005), score: 0.79 ST6GAL2ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 (ENSG00000144057), score: 0.66 ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (ENSG00000117069), score: 0.84 STOML1stomatin (EPB72)-like 1 (ENSG00000067221), score: 0.71 SULT4A1sulfotransferase family 4A, member 1 (ENSG00000130540), score: 0.66 SV2Bsynaptic vesicle glycoprotein 2B (ENSG00000185518), score: 0.66 SYF2SYF2 homolog, RNA splicing factor (S. cerevisiae) (ENSG00000117614), score: -0.6 SYN2synapsin II (ENSG00000157152), score: 0.67 SYT17synaptotagmin XVII (ENSG00000103528), score: 0.78 TBC1D23TBC1 domain family, member 23 (ENSG00000036054), score: -0.64 TDRD3tudor domain containing 3 (ENSG00000083544), score: -0.63 TGFBR1transforming growth factor, beta receptor 1 (ENSG00000106799), score: -0.76 THOC2THO complex 2 (ENSG00000125676), score: -0.65 TMEM179transmembrane protein 179 (ENSG00000189203), score: 0.79 TMEM59Ltransmembrane protein 59-like (ENSG00000105696), score: 0.73 TNRtenascin R (restrictin, janusin) (ENSG00000116147), score: 0.65 TOXthymocyte selection-associated high mobility group box (ENSG00000198846), score: 0.73 TP53INP1tumor protein p53 inducible nuclear protein 1 (ENSG00000164938), score: -0.66 TPPPtubulin polymerization promoting protein (ENSG00000171368), score: 0.72 TRAF7TNF receptor-associated factor 7 (ENSG00000131653), score: 0.66 TRIP4thyroid hormone receptor interactor 4 (ENSG00000103671), score: -0.65 TRPM2transient receptor potential cation channel, subfamily M, member 2 (ENSG00000142185), score: 0.71 TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: -0.71 TSHZ3teashirt zinc finger homeobox 3 (ENSG00000121297), score: 0.7 TTC27tetratricopeptide repeat domain 27 (ENSG00000018699), score: -0.6 UNC5Aunc-5 homolog A (C. elegans) (ENSG00000113763), score: 0.79 UNC5Dunc-5 homolog D (C. elegans) (ENSG00000156687), score: 0.67 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: -0.58 USP3ubiquitin specific peptidase 3 (ENSG00000140455), score: -0.61 VIPvasoactive intestinal peptide (ENSG00000146469), score: 0.7 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: -0.61 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: -0.72 VSTM2AV-set and transmembrane domain containing 2A (ENSG00000170419), score: 0.72 VWC2Lvon Willebrand factor C domain-containing protein 2-like (ENSG00000174453), score: 0.73 WDR37WD repeat domain 37 (ENSG00000047056), score: 0.75 WDR7WD repeat domain 7 (ENSG00000091157), score: 0.71 WEE1WEE1 homolog (S. pombe) (ENSG00000166483), score: -0.69 WNT7Awingless-type MMTV integration site family, member 7A (ENSG00000154764), score: 0.77 WSB2WD repeat and SOCS box-containing 2 (ENSG00000176871), score: 0.66 XRN15'-3' exoribonuclease 1 (ENSG00000114127), score: -0.65 XRN25'-3' exoribonuclease 2 (ENSG00000088930), score: -0.61 XYLT1xylosyltransferase I (ENSG00000103489), score: 0.75 YWHAHtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide (ENSG00000128245), score: 0.69 ZDHHC22zinc finger, DHHC-type containing 22 (ENSG00000177108), score: 0.85 ZER1zer-1 homolog (C. elegans) (ENSG00000160445), score: 0.72 ZFAND6zinc finger, AN1-type domain 6 (ENSG00000086666), score: -0.77 ZMAT4zinc finger, matrin type 4 (ENSG00000165061), score: 0.67 ZNF365zinc finger protein 365 (ENSG00000138311), score: 0.68
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_br_m_ca1 | ggo | br | m | _ |
| mml_br_f_ca1 | mml | br | f | _ |
| ppy_br_f_ca1 | ppy | br | f | _ |
| mml_br_m2_ca1 | mml | br | m | 2 |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| ppy_br_m_ca1 | ppy | br | m | _ |
| hsa_br_m7_ca1 | hsa | br | m | 7 |