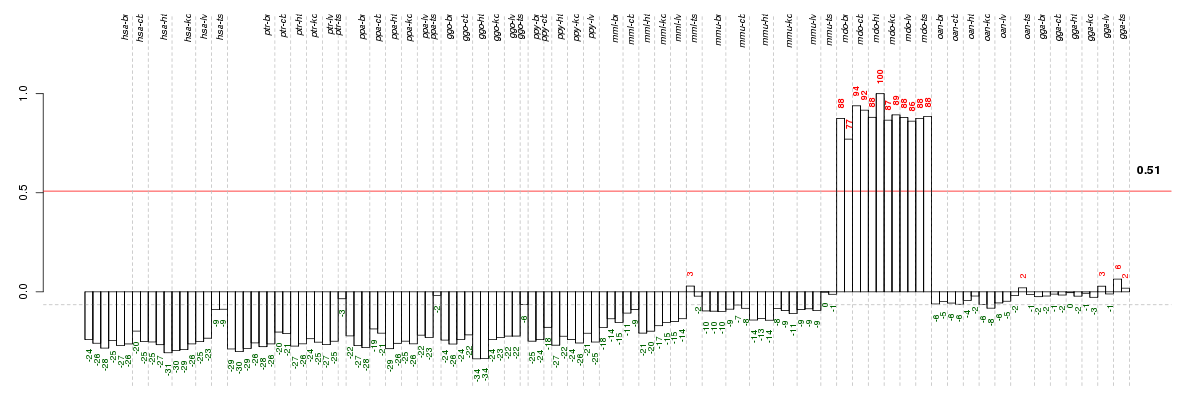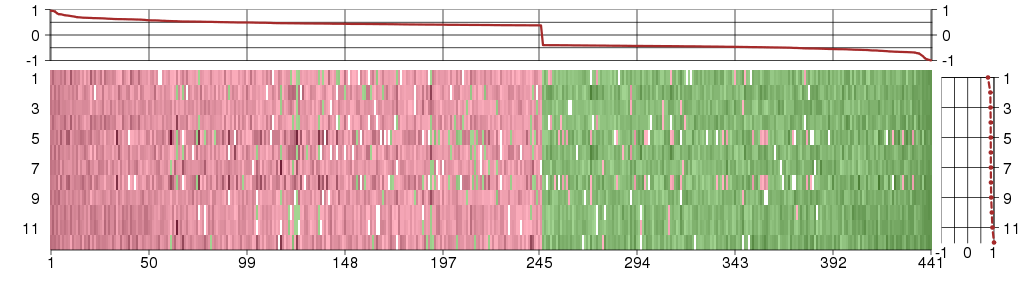

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
RNA processing
Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
RNA processing
Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.

AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: -0.61 AATFapoptosis antagonizing transcription factor (ENSG00000108270), score: -0.57 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: 0.41 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (ENSG00000160179), score: -0.51 ACANaggrecan (ENSG00000157766), score: 0.46 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: 0.58 ACBD6acyl-CoA binding domain containing 6 (ENSG00000135847), score: -0.49 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.52 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.38 ACTL6Aactin-like 6A (ENSG00000136518), score: 0.44 ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.68 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.63 AHSA2AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) (ENSG00000173209), score: -0.62 AIREautoimmune regulator (ENSG00000160224), score: 0.52 ALKBH1alkB, alkylation repair homolog 1 (E. coli) (ENSG00000100601), score: 0.52 ALKBH2alkB, alkylation repair homolog 2 (E. coli) (ENSG00000189046), score: 0.62 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.61 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.48 ANKRD10ankyrin repeat domain 10 (ENSG00000088448), score: -0.41 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.5 ANKRD16ankyrin repeat domain 16 (ENSG00000134461), score: -0.43 ANKRD42ankyrin repeat domain 42 (ENSG00000137494), score: -0.4 APAF1apoptotic peptidase activating factor 1 (ENSG00000120868), score: 0.48 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: 0.43 ARFRP1ADP-ribosylation factor related protein 1 (ENSG00000101246), score: -0.45 ARHGAP24Rho GTPase activating protein 24 (ENSG00000138639), score: -0.4 ARHGAP40Rho GTPase activating protein 40 (ENSG00000124143), score: 0.43 ARHGDIGRho GDP dissociation inhibitor (GDI) gamma (ENSG00000167930), score: -0.44 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 0.41 ARHGEF7Rho guanine nucleotide exchange factor (GEF) 7 (ENSG00000102606), score: -0.42 ARL9ADP-ribosylation factor-like 9 (ENSG00000196503), score: 0.41 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.64 ASB1ankyrin repeat and SOCS box-containing 1 (ENSG00000065802), score: 0.61 ASXL2additional sex combs like 2 (Drosophila) (ENSG00000143970), score: 0.42 ATG16L1ATG16 autophagy related 16-like 1 (S. cerevisiae) (ENSG00000085978), score: 0.61 ATG4BATG4 autophagy related 4 homolog B (S. cerevisiae) (ENSG00000168397), score: -0.43 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: 0.66 ATMINATM interactor (ENSG00000166454), score: -0.47 ATP13A2ATPase type 13A2 (ENSG00000159363), score: -0.47 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.6 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.38 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.67 ATXN2ataxin 2 (ENSG00000204842), score: 0.44 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: -0.54 BACE2beta-site APP-cleaving enzyme 2 (ENSG00000182240), score: -0.43 BCL2L13BCL2-like 13 (apoptosis facilitator) (ENSG00000099968), score: -0.56 BCL7AB-cell CLL/lymphoma 7A (ENSG00000110987), score: 0.39 BLVRAbiliverdin reductase A (ENSG00000106605), score: -0.68 BMP15bone morphogenetic protein 15 (ENSG00000130385), score: 0.38 BRF2BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like (ENSG00000104221), score: -0.43 BRPF1bromodomain and PHD finger containing, 1 (ENSG00000156983), score: 0.43 BST1bone marrow stromal cell antigen 1 (ENSG00000109743), score: -0.41 BTKBruton agammaglobulinemia tyrosine kinase (ENSG00000010671), score: 0.4 C10orf137chromosome 10 open reading frame 137 (ENSG00000107938), score: 0.49 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.67 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.66 C12orf23chromosome 12 open reading frame 23 (ENSG00000151135), score: 0.38 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.6 C13orf31chromosome 13 open reading frame 31 (ENSG00000179630), score: 0.74 C14orf101chromosome 14 open reading frame 101 (ENSG00000070269), score: -0.55 C15orf29chromosome 15 open reading frame 29 (ENSG00000134152), score: -0.5 C15orf60chromosome 15 open reading frame 60 (ENSG00000183324), score: 0.49 C17orf39chromosome 17 open reading frame 39 (ENSG00000141034), score: 0.38 C17orf71chromosome 17 open reading frame 71 (ENSG00000167447), score: 0.53 C17orf75chromosome 17 open reading frame 75 (ENSG00000108666), score: 0.4 C1orf103chromosome 1 open reading frame 103 (ENSG00000121931), score: -0.45 C1orf123chromosome 1 open reading frame 123 (ENSG00000162384), score: 0.61 C1orf159chromosome 1 open reading frame 159 (ENSG00000131591), score: 0.43 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.63 C22orf13chromosome 22 open reading frame 13 (ENSG00000138867), score: 0.42 C2CD2C2 calcium-dependent domain containing 2 (ENSG00000157617), score: -0.54 C3orf23chromosome 3 open reading frame 23 (ENSG00000179152), score: 0.54 C3orf39chromosome 3 open reading frame 39 (ENSG00000144647), score: 0.69 C3orf59chromosome 3 open reading frame 59 (ENSG00000180611), score: 0.51 C5orf24chromosome 5 open reading frame 24 (ENSG00000181904), score: -0.43 C6orf57chromosome 6 open reading frame 57 (ENSG00000154079), score: 0.47 C6orf64chromosome 6 open reading frame 64 (ENSG00000112167), score: -0.45 C6orf89chromosome 6 open reading frame 89 (ENSG00000198663), score: 0.5 C9orf102chromosome 9 open reading frame 102 (ENSG00000182150), score: -0.48 C9orf103chromosome 9 open reading frame 103 (ENSG00000148057), score: -0.49 CA13carbonic anhydrase XIII (ENSG00000185015), score: 0.53 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.44 CARS2cysteinyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000134905), score: -0.41 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (ENSG00000078699), score: 0.4 CCDC6coiled-coil domain containing 6 (ENSG00000108091), score: 0.41 CCDC82coiled-coil domain containing 82 (ENSG00000149231), score: -0.53 CCM2cerebral cavernous malformation 2 (ENSG00000136280), score: -0.47 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.65 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: -0.64 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.79 CDK10cyclin-dependent kinase 10 (ENSG00000185324), score: -0.56 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (ENSG00000115816), score: -0.4 CENPNcentromere protein N (ENSG00000166451), score: -0.41 CENPTcentromere protein T (ENSG00000102901), score: -0.43 CFDP1craniofacial development protein 1 (ENSG00000153774), score: -0.48 CHD1Lchromodomain helicase DNA binding protein 1-like (ENSG00000131778), score: -0.51 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: -0.48 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.48 CLIC3chloride intracellular channel 3 (ENSG00000169583), score: 0.45 CLTBclathrin, light chain B (ENSG00000175416), score: -0.56 CNTFciliary neurotrophic factor (ENSG00000186660), score: 0.55 COBRA1cofactor of BRCA1 (ENSG00000188986), score: -0.43 COG7component of oligomeric golgi complex 7 (ENSG00000168434), score: -0.44 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.92 CPSF6cleavage and polyadenylation specific factor 6, 68kDa (ENSG00000111605), score: 0.46 CRTAPcartilage associated protein (ENSG00000170275), score: -0.47 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.49 CSTF1cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa (ENSG00000101138), score: -0.43 CYLDcylindromatosis (turban tumor syndrome) (ENSG00000083799), score: -0.6 CYTBcytochrome b (ENSG00000198727), score: -1 CYTH1cytohesin 1 (ENSG00000108669), score: -0.69 DCTDdCMP deaminase (ENSG00000129187), score: -0.45 DCTN5dynactin 5 (p25) (ENSG00000166847), score: -0.59 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (ENSG00000109184), score: -0.44 DDX1DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (ENSG00000079785), score: 0.69 DDX24DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 (ENSG00000089737), score: -0.42 DDX41DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (ENSG00000183258), score: -0.55 DDX47DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (ENSG00000213782), score: -0.43 DDX51DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 (ENSG00000185163), score: -0.54 DENND5BDENN/MADD domain containing 5B (ENSG00000170456), score: 0.44 DERL3Der1-like domain family, member 3 (ENSG00000099958), score: 0.49 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: -0.66 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: 0.42 DNM1Ldynamin 1-like (ENSG00000087470), score: 0.48 DPH5DPH5 homolog (S. cerevisiae) (ENSG00000117543), score: 0.51 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (ENSG00000105865), score: -0.49 DUSP4dual specificity phosphatase 4 (ENSG00000120875), score: 0.41 ECDecdysoneless homolog (Drosophila) (ENSG00000122882), score: 0.62 EEA1early endosome antigen 1 (ENSG00000102189), score: 0.46 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (ENSG00000172071), score: 0.43 EIF2B1eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa (ENSG00000111361), score: -0.41 EIF2C2eukaryotic translation initiation factor 2C, 2 (ENSG00000123908), score: -0.41 ELMO3engulfment and cell motility 3 (ENSG00000102890), score: 0.41 EME1essential meiotic endonuclease 1 homolog 1 (S. pombe) (ENSG00000154920), score: 0.45 EPB41L4Aerythrocyte membrane protein band 4.1 like 4A (ENSG00000129595), score: -0.45 ERCC5excision repair cross-complementing rodent repair deficiency, complementation group 5 (ENSG00000134899), score: -0.4 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.66 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.61 ETAA1Ewing tumor-associated antigen 1 (ENSG00000143971), score: 0.41 EVI5ecotropic viral integration site 5 (ENSG00000067208), score: 0.38 EXD2exonuclease 3'-5' domain containing 2 (ENSG00000081177), score: -0.53 EXOC3exocyst complex component 3 (ENSG00000180104), score: -0.57 EXOSC9exosome component 9 (ENSG00000123737), score: 0.56 FAM13Cfamily with sequence similarity 13, member C (ENSG00000148541), score: 0.4 FAM168Bfamily with sequence similarity 168, member B (ENSG00000152102), score: -0.47 FAM172Afamily with sequence similarity 172, member A (ENSG00000113391), score: 0.47 FAM20Bfamily with sequence similarity 20, member B (ENSG00000116199), score: 0.52 FAM3Bfamily with sequence similarity 3, member B (ENSG00000183844), score: 0.38 FAM40Bfamily with sequence similarity 40, member B (ENSG00000128578), score: 0.43 FAM73Afamily with sequence similarity 73, member A (ENSG00000180488), score: 0.62 FAM82A2family with sequence similarity 82, member A2 (ENSG00000137824), score: 0.62 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: -0.45 FBXO15F-box protein 15 (ENSG00000141665), score: 0.53 FBXO22F-box protein 22 (ENSG00000167196), score: 0.64 FBXO3F-box protein 3 (ENSG00000110429), score: 0.62 FGF4fibroblast growth factor 4 (ENSG00000075388), score: 0.46 FKBP10FK506 binding protein 10, 65 kDa (ENSG00000141756), score: -0.42 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: 0.45 FLVCR1feline leukemia virus subgroup C cellular receptor 1 (ENSG00000162769), score: 0.49 FN3KRPfructosamine 3 kinase related protein (ENSG00000141560), score: -0.41 FOXN1forkhead box N1 (ENSG00000109101), score: 0.55 FRA10AC1fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 (ENSG00000148690), score: -0.58 FRZBfrizzled-related protein (ENSG00000162998), score: -0.43 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.85 GARSglycyl-tRNA synthetase (ENSG00000106105), score: 0.66 GBF1golgi brefeldin A resistant guanine nucleotide exchange factor 1 (ENSG00000107862), score: 0.41 GBGT1globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 (ENSG00000148288), score: 0.49 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.52 GEMIN5gem (nuclear organelle) associated protein 5 (ENSG00000082516), score: 0.5 GFM2G elongation factor, mitochondrial 2 (ENSG00000164347), score: -0.48 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: 0.46 GLMNglomulin, FKBP associated protein (ENSG00000174842), score: 0.43 GLT8D1glycosyltransferase 8 domain containing 1 (ENSG00000016864), score: -0.49 GMNNgeminin, DNA replication inhibitor (ENSG00000112312), score: -0.43 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: -0.46 GNPTGN-acetylglucosamine-1-phosphate transferase, gamma subunit (ENSG00000090581), score: -0.42 GPBP1L1GC-rich promoter binding protein 1-like 1 (ENSG00000159592), score: 0.38 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.65 GPX8glutathione peroxidase 8 (putative) (ENSG00000164294), score: 0.38 GRAMD2GRAM domain containing 2 (ENSG00000175318), score: 0.41 GRAMD3GRAM domain containing 3 (ENSG00000155324), score: -0.43 GRHL1grainyhead-like 1 (Drosophila) (ENSG00000134317), score: -0.41 GTF3Ageneral transcription factor IIIA (ENSG00000122034), score: -0.58 HAGHhydroxyacylglutathione hydrolase (ENSG00000063854), score: -0.46 HEATR6HEAT repeat containing 6 (ENSG00000068097), score: 0.45 HELLShelicase, lymphoid-specific (ENSG00000119969), score: 0.47 HNRNPA0heterogeneous nuclear ribonucleoprotein A0 (ENSG00000177733), score: 0.39 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (ENSG00000153187), score: 0.56 HPRT1hypoxanthine phosphoribosyltransferase 1 (ENSG00000165704), score: 0.48 HSF1heat shock transcription factor 1 (ENSG00000185122), score: -0.46 IDEinsulin-degrading enzyme (ENSG00000119912), score: 0.4 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.72 IKBIPIKBKB interacting protein (ENSG00000166130), score: 0.61 IL10RBinterleukin 10 receptor, beta (ENSG00000243646), score: -0.47 INVSinversin (ENSG00000119509), score: -0.59 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.94 IRAK1BP1interleukin-1 receptor-associated kinase 1 binding protein 1 (ENSG00000146243), score: 0.51 KBTBD4kelch repeat and BTB (POZ) domain containing 4 (ENSG00000123444), score: -0.44 KBTBD8kelch repeat and BTB (POZ) domain containing 8 (ENSG00000163376), score: 0.57 KCTD5potassium channel tetramerisation domain containing 5 (ENSG00000167977), score: -0.55 KCTD6potassium channel tetramerisation domain containing 6 (ENSG00000168301), score: 0.44 KDM3Alysine (K)-specific demethylase 3A (ENSG00000115548), score: 0.43 KIAA0090KIAA0090 (ENSG00000127463), score: 0.44 KIAA0146KIAA0146 (ENSG00000164808), score: -0.54 KIAA0174KIAA0174 (ENSG00000182149), score: -0.62 KIAA1432KIAA1432 (ENSG00000107036), score: 0.53 KIAA1712KIAA1712 (ENSG00000164118), score: 0.45 KIAA1841KIAA1841 (ENSG00000162929), score: 0.39 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.41 LAMP2lysosomal-associated membrane protein 2 (ENSG00000005893), score: -0.4 LCA5Leber congenital amaurosis 5 (ENSG00000135338), score: 0.43 LEPRE1leucine proline-enriched proteoglycan (leprecan) 1 (ENSG00000117385), score: 0.56 LOC100134387similar to DEAH (Asp-Glu-Ala-His) box polypeptide 40 (ENSG00000108406), score: -0.46 LOC100288211PPPDE peptidase domain containing 1 pseudogene (ENSG00000121644), score: 0.48 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: 0.53 LRATlecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) (ENSG00000121207), score: 0.46 LRRC40leucine rich repeat containing 40 (ENSG00000066557), score: 0.39 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: -0.53 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (ENSG00000197063), score: -0.45 MARCH2membrane-associated ring finger (C3HC4) 2 (ENSG00000099785), score: 0.38 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (ENSG00000082212), score: -0.48 MED1mediator complex subunit 1 (ENSG00000125686), score: 0.57 MED13mediator complex subunit 13 (ENSG00000108510), score: 0.4 MEMO1mediator of cell motility 1 (ENSG00000162961), score: -0.66 MESDC1mesoderm development candidate 1 (ENSG00000140406), score: 0.42 METRNLmeteorin, glial cell differentiation regulator-like (ENSG00000176845), score: -0.6 MFN1mitofusin 1 (ENSG00000171109), score: 0.45 MGAT4Amannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A (ENSG00000071073), score: -0.45 MGST2microsomal glutathione S-transferase 2 (ENSG00000085871), score: -0.46 MID2midline 2 (ENSG00000080561), score: 0.52 MPP6membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (ENSG00000105926), score: 0.38 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.65 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: -0.43 MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.38 MTHFD2Lmethylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (ENSG00000163738), score: 0.4 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: -0.57 MTMR14myotubularin related protein 14 (ENSG00000163719), score: 0.46 MTMR3myotubularin related protein 3 (ENSG00000100330), score: 0.45 MTRR5-methyltetrahydrofolate-homocysteine methyltransferase reductase (ENSG00000124275), score: -0.44 MYH10myosin, heavy chain 10, non-muscle (ENSG00000133026), score: -0.41 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.7 NADSYN1NAD synthetase 1 (ENSG00000172890), score: 0.43 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.51 NARFnuclear prelamin A recognition factor (ENSG00000141562), score: -0.67 NARG2NMDA receptor regulated 2 (ENSG00000128915), score: -0.51 ND2MTND2 (ENSG00000198763), score: -0.95 ND4NADH dehydrogenase, subunit 4 (complex I) (ENSG00000198886), score: -0.45 ND6NADH dehydrogenase, subunit 6 (complex I) (ENSG00000198695), score: -0.62 NDUFA6NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa (ENSG00000184983), score: 0.43 NEK8NIMA (never in mitosis gene a)- related kinase 8 (ENSG00000160602), score: 0.41 NFIAnuclear factor I/A (ENSG00000162599), score: 0.4 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: 0.61 NINninein (GSK3B interacting protein) (ENSG00000100503), score: -0.42 NMT2N-myristoyltransferase 2 (ENSG00000152465), score: -0.47 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.92 NPLOC4nuclear protein localization 4 homolog (S. cerevisiae) (ENSG00000182446), score: -0.45 NRBF2nuclear receptor binding factor 2 (ENSG00000148572), score: 0.45 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.72 NT5DC35'-nucleotidase domain containing 3 (ENSG00000111696), score: 0.4 NUDCD3NudC domain containing 3 (ENSG00000015676), score: -0.47 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.47 NUDT3nudix (nucleoside diphosphate linked moiety X)-type motif 3 (ENSG00000112664), score: -0.42 NUF2NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000143228), score: 0.4 NUP153nucleoporin 153kDa (ENSG00000124789), score: 0.39 NUP54nucleoporin 54kDa (ENSG00000138750), score: 0.49 NUP85nucleoporin 85kDa (ENSG00000125450), score: 0.49 OCIAD1OCIA domain containing 1 (ENSG00000109180), score: -0.49 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: -0.52 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.41 OTOL1otolin 1 homolog (zebrafish) (ENSG00000182447), score: 0.54 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: -0.49 PAPD4PAP associated domain containing 4 (ENSG00000164329), score: 0.46 PCID2PCI domain containing 2 (ENSG00000126226), score: -0.51 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.64 PDE9Aphosphodiesterase 9A (ENSG00000160191), score: -0.46 PDS5APDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) (ENSG00000121892), score: -0.42 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.53 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (ENSG00000170525), score: -0.46 PGRprogesterone receptor (ENSG00000082175), score: 0.66 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.62 PHF21APHD finger protein 21A (ENSG00000135365), score: 0.59 PHF5APHD finger protein 5A (ENSG00000100410), score: 0.57 PHKA2phosphorylase kinase, alpha 2 (liver) (ENSG00000044446), score: 0.39 PHTF1putative homeodomain transcription factor 1 (ENSG00000116793), score: 0.45 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.63 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.73 PIGSphosphatidylinositol glycan anchor biosynthesis, class S (ENSG00000087111), score: 0.65 PLDNpallidin homolog (mouse) (ENSG00000104164), score: -0.5 PNPLA6patatin-like phospholipase domain containing 6 (ENSG00000032444), score: 0.4 PODNpodocan (ENSG00000174348), score: 0.57 POLR2Cpolymerase (RNA) II (DNA directed) polypeptide C, 33kDa (ENSG00000102978), score: -0.41 POLR3Apolymerase (RNA) III (DNA directed) polypeptide A, 155kDa (ENSG00000148606), score: -0.4 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (ENSG00000161980), score: 0.41 PPIP5K2diphosphoinositol pentakisphosphate kinase 2 (ENSG00000145725), score: 0.47 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta (ENSG00000175470), score: 0.41 PPTC7PTC7 protein phosphatase homolog (S. cerevisiae) (ENSG00000196850), score: 0.42 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.56 PRKACBprotein kinase, cAMP-dependent, catalytic, beta (ENSG00000142875), score: -0.56 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.45 PSEN1presenilin 1 (ENSG00000080815), score: -0.47 PSEN2presenilin 2 (Alzheimer disease 4) (ENSG00000143801), score: 0.38 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (ENSG00000159377), score: -0.44 PSMG1proteasome (prosome, macropain) assembly chaperone 1 (ENSG00000183527), score: 0.43 PTCH1patched 1 (ENSG00000185920), score: -0.41 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: -0.4 PTRH2peptidyl-tRNA hydrolase 2 (ENSG00000141378), score: -0.53 PUS10pseudouridylate synthase 10 (ENSG00000162927), score: 0.38 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.76 QSER1glutamine and serine rich 1 (ENSG00000060749), score: 0.45 QSOX1quiescin Q6 sulfhydryl oxidase 1 (ENSG00000116260), score: -0.43 RABEPKRab9 effector protein with kelch motifs (ENSG00000136933), score: -0.5 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.5 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.48 RALAv-ral simian leukemia viral oncogene homolog A (ras related) (ENSG00000006451), score: 0.39 RAP2CRAP2C, member of RAS oncogene family (ENSG00000123728), score: 0.39 RASL11ARAS-like, family 11, member A (ENSG00000122035), score: -0.44 RBM6RNA binding motif protein 6 (ENSG00000004534), score: -0.46 RBPJLrecombination signal binding protein for immunoglobulin kappa J region-like (ENSG00000124232), score: 0.53 RC3H1ring finger and CCCH-type zinc finger domains 1 (ENSG00000135870), score: 0.4 RCAN1regulator of calcineurin 1 (ENSG00000159200), score: -0.49 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.4 RFXANKregulatory factor X-associated ankyrin-containing protein (ENSG00000064490), score: 0.46 RG9MTD1RNA (guanine-9-) methyltransferase domain containing 1 (ENSG00000174173), score: -0.58 RGS2regulator of G-protein signaling 2, 24kDa (ENSG00000116741), score: -0.44 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.77 RNF145ring finger protein 145 (ENSG00000145860), score: -0.4 RNF43ring finger protein 43 (ENSG00000108375), score: 0.4 RPGRIP1LRPGRIP1-like (ENSG00000103494), score: 0.39 RPS6KA2ribosomal protein S6 kinase, 90kDa, polypeptide 2 (ENSG00000071242), score: -0.4 RPS6KA5ribosomal protein S6 kinase, 90kDa, polypeptide 5 (ENSG00000100784), score: 0.41 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.68 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.53 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.41 SAP30LSAP30-like (ENSG00000164576), score: 0.66 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.55 SEH1LSEH1-like (S. cerevisiae) (ENSG00000085415), score: 0.45 SELSselenoprotein S (ENSG00000131871), score: -0.43 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.56 SFRS8splicing factor, arginine/serine-rich 8 (suppressor-of-white-apricot homolog, Drosophila) (ENSG00000061936), score: 0.66 SGSM2small G protein signaling modulator 2 (ENSG00000141258), score: -0.4 SIN3BSIN3 homolog B, transcription regulator (yeast) (ENSG00000127511), score: -0.49 SIRT1sirtuin 1 (ENSG00000096717), score: 0.5 SKILSKI-like oncogene (ENSG00000136603), score: 0.48 SLC15A4solute carrier family 15, member 4 (ENSG00000139370), score: 0.43 SLC25A38solute carrier family 25, member 38 (ENSG00000144659), score: -0.45 SLC27A1solute carrier family 27 (fatty acid transporter), member 1 (ENSG00000130304), score: 0.4 SLC30A9solute carrier family 30 (zinc transporter), member 9 (ENSG00000014824), score: -0.44 SLC35B3solute carrier family 35, member B3 (ENSG00000124786), score: -0.48 SLC38A1solute carrier family 38, member 1 (ENSG00000111371), score: -0.42 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.95 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (ENSG00000100335), score: 0.45 SMUG1single-strand-selective monofunctional uracil-DNA glycosylase 1 (ENSG00000123415), score: -0.44 SNRKSNF related kinase (ENSG00000163788), score: 0.54 SNRPA1small nuclear ribonucleoprotein polypeptide A' (ENSG00000131876), score: -0.4 SNX13sorting nexin 13 (ENSG00000071189), score: 0.67 SNX24sorting nexin 24 (ENSG00000064652), score: 0.43 SNX8sorting nexin 8 (ENSG00000106266), score: -0.46 SPG20spastic paraplegia 20 (Troyer syndrome) (ENSG00000133104), score: -0.59 SRPX2sushi-repeat-containing protein, X-linked 2 (ENSG00000102359), score: 0.58 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: 0.44 STAMBPSTAM binding protein (ENSG00000124356), score: 0.52 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (ENSG00000040341), score: -0.53 STILSCL/TAL1 interrupting locus (ENSG00000123473), score: 0.48 STK10serine/threonine kinase 10 (ENSG00000072786), score: -0.42 STRBPspermatid perinuclear RNA binding protein (ENSG00000165209), score: 0.42 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.68 STX16syntaxin 16 (ENSG00000124222), score: -0.52 STX8syntaxin 8 (ENSG00000170310), score: -0.58 SUPT5Hsuppressor of Ty 5 homolog (S. cerevisiae) (ENSG00000196235), score: 0.39 SUSD1sushi domain containing 1 (ENSG00000106868), score: -0.43 SYF2SYF2 homolog, RNA splicing factor (S. cerevisiae) (ENSG00000117614), score: -0.42 SYT12synaptotagmin XII (ENSG00000173227), score: 0.39 TAB2TGF-beta activated kinase 1/MAP3K7 binding protein 2 (ENSG00000055208), score: -0.44 TARSthreonyl-tRNA synthetase (ENSG00000113407), score: -0.41 TCF3transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) (ENSG00000071564), score: 0.5 TCP11L2t-complex 11 (mouse)-like 2 (ENSG00000166046), score: 0.43 TECTBtectorin beta (ENSG00000119913), score: 0.48 TERTtelomerase reverse transcriptase (ENSG00000164362), score: 0.5 TEX264testis expressed 264 (ENSG00000164081), score: 0.38 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.78 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (ENSG00000151923), score: 0.49 TMCO3transmembrane and coiled-coil domains 3 (ENSG00000150403), score: -0.4 TMCO6transmembrane and coiled-coil domains 6 (ENSG00000113119), score: 0.57 TMEM100transmembrane protein 100 (ENSG00000166292), score: 0.4 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.43 TMEM138transmembrane protein 138 (ENSG00000149483), score: 0.53 TMEM18transmembrane protein 18 (ENSG00000151353), score: -0.68 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.81 TMEM87Atransmembrane protein 87A (ENSG00000103978), score: 0.38 TNFSF13Btumor necrosis factor (ligand) superfamily, member 13b (ENSG00000102524), score: -0.42 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.77 TRAF1TNF receptor-associated factor 1 (ENSG00000056558), score: -0.48 TRAF3TNF receptor-associated factor 3 (ENSG00000131323), score: -0.44 TRIP12thyroid hormone receptor interactor 12 (ENSG00000153827), score: 0.41 TSC22D2TSC22 domain family, member 2 (ENSG00000196428), score: 0.49 TSNtranslin (ENSG00000211460), score: -0.67 TSPAN13tetraspanin 13 (ENSG00000106537), score: 0.38 TSPAN9tetraspanin 9 (ENSG00000011105), score: 0.39 TTF2transcription termination factor, RNA polymerase II (ENSG00000116830), score: 0.42 TTLL1tubulin tyrosine ligase-like family, member 1 (ENSG00000100271), score: 0.43 TTLL9tubulin tyrosine ligase-like family, member 9 (ENSG00000131044), score: 0.39 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.53 TUFT1tuftelin 1 (ENSG00000143367), score: 0.44 TUSC3tumor suppressor candidate 3 (ENSG00000104723), score: 0.44 TXNDC12thioredoxin domain containing 12 (endoplasmic reticulum) (ENSG00000117862), score: 0.46 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.83 UBTD2ubiquitin domain containing 2 (ENSG00000168246), score: 0.41 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.55 UCK1uridine-cytidine kinase 1 (ENSG00000130717), score: 0.42 UNC50unc-50 homolog (C. elegans) (ENSG00000115446), score: -0.61 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: 0.63 UTP11LUTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) (ENSG00000183520), score: 0.42 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.68 VPS33Bvacuolar protein sorting 33 homolog B (yeast) (ENSG00000184056), score: 0.45 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (ENSG00000139722), score: 0.38 VPS39vacuolar protein sorting 39 homolog (S. cerevisiae) (ENSG00000166887), score: -0.42 WBSCR22Williams Beuren syndrome chromosome region 22 (ENSG00000071462), score: -0.4 WDR43WD repeat domain 43 (ENSG00000163811), score: 0.43 WDR6WD repeat domain 6 (ENSG00000178252), score: -0.45 WFDC1WAP four-disulfide core domain 1 (ENSG00000103175), score: 0.38 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: 0.53 YAF2YY1 associated factor 2 (ENSG00000015153), score: -0.4 ZBED4zinc finger, BED-type containing 4 (ENSG00000100426), score: 0.44 ZBTB11zinc finger and BTB domain containing 11 (ENSG00000066422), score: -0.45 ZBTB8Azinc finger and BTB domain containing 8A (ENSG00000160062), score: -0.44 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.97 ZCCHC8zinc finger, CCHC domain containing 8 (ENSG00000033030), score: 0.44 ZDHHC21zinc finger, DHHC-type containing 21 (ENSG00000175893), score: -0.46 ZDHHC6zinc finger, DHHC-type containing 6 (ENSG00000023041), score: -0.41 ZFP161zinc finger protein 161 homolog (mouse) (ENSG00000198081), score: -0.42 ZFYVE27zinc finger, FYVE domain containing 27 (ENSG00000155256), score: 0.43 ZGPATzinc finger, CCCH-type with G patch domain (ENSG00000197114), score: 0.4 ZMYND10zinc finger, MYND-type containing 10 (ENSG00000004838), score: 0.45 ZNF318zinc finger protein 318 (ENSG00000171467), score: 0.39 ZNF346zinc finger protein 346 (ENSG00000113761), score: -0.51 ZNF385Czinc finger protein 385C (ENSG00000187595), score: 0.42 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.81 ZNF704zinc finger protein 704 (ENSG00000164684), score: -0.4 ZNF800zinc finger protein 800 (ENSG00000048405), score: 0.52 ZRANB3zinc finger, RAN-binding domain containing 3 (ENSG00000121988), score: -0.42 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.72
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_br_f_ca1 | mdo | br | f | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |