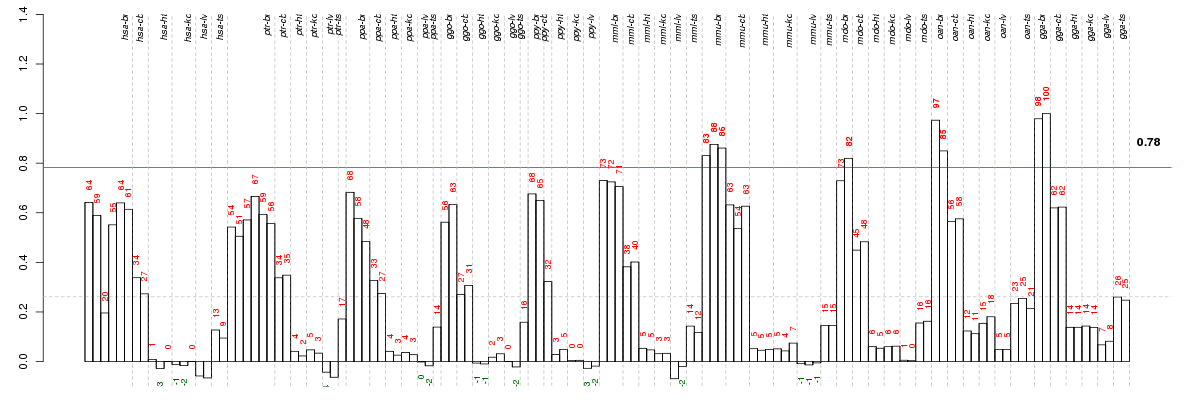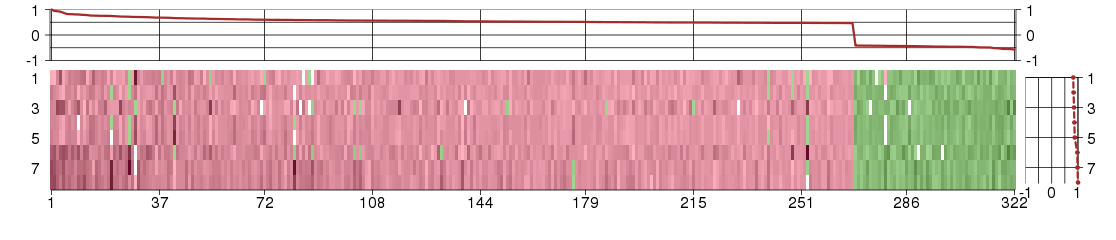

Under-expression is coded with green,
over-expression with red color.

cell morphogenesis
The developmental process by which the size or shape of a cell is generated and organized. Morphogenesis pertains to the creation of form.
cell morphogenesis involved in differentiation
The change in form (cell shape and size) that occurs when relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
purine nucleotide metabolic process
The chemical reactions and pathways involving a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
purine nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
cAMP biosynthetic process
The chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
nucleoside phosphate metabolic process
The chemical reactions and pathways involving any phosphorylated nucleoside.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cation transport
The directed movement of cations, atoms or small molecules with a net positive charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
calcium ion transport
The directed movement of calcium (Ca) ions into, out of, within or between cells by means of some external agent such as a transporter or pore.
metal ion transport
The directed movement of metal ions, any metal ion with an electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
G-protein coupled receptor protein signaling pathway
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand.
G-protein signaling, coupled to cyclic nucleotide second messenger
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand, followed by modulation of a nucleotide cyclase activity and a subsequent change in the concentration of a cyclic nucleotide.
G-protein signaling, coupled to cAMP nucleotide second messenger
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand, followed by modulation of adenylyl cyclase activity and a subsequent change in the concentration of cyclic AMP.
negative regulation of adenylate cyclase activity
Any process that stops, prevents or reduces the frequency, rate or extent of adenylate cyclase activity.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
axonogenesis
Generation of a long process of a neuron, that carries efferent (outgoing) action potentials from the cell body towards target cells.
behavior
The specific actions or reactions of an organism in response to external or internal stimuli. Patterned activity of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions.
learning or memory
The acquisition and processing of information and/or the storage and retrieval of this information over time.
memory
The activities involved in the mental information processing system that receives (registers), modifies, stores, and retrieves informational stimuli. The main stages involved in the formation and retrieval of memory are encoding (processing of received information by acquisition), storage (building a permanent record of received information as a result of consolidation) and retrieval (calling back the stored information and use it in a suitable way to execute a given task).
long-term memory
The activities involved in the mental information processing system that deals with the storage, retrieval and modification of information a long time (typically weeks, months or years) after receiving that information. This type of memory is typically dependent on gene transcription regulated by second messenger activation.
cell recognition
The process by which a cell in a multicellular organism interprets its surroundings.
neuron recognition
The process by which a neuronal cell in a multicellular organism interprets its surroundings.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cAMP metabolic process
The chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
nucleotide metabolic process
The chemical reactions and pathways involving a nucleotide, a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic nucleotides (nucleoside cyclic phosphates).
nucleoside monophosphate metabolic process
The chemical reactions and pathways involving a nucleoside monophosphate, a glycosamine consisting of a base linked to a deoxyribose or ribose sugar esterified with phosphate on its glycose moiety.
nucleoside monophosphate biosynthetic process
The chemical reactions and pathways resulting in the formation of a nucleoside monophosphate, a glycosamine consisting of a base linked to a deoxyribose or ribose sugar esterified with phosphate on its glycose moiety.
nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleotides, any nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic-nucleotides (nucleoside cyclic phosphates).
cyclic nucleotide metabolic process
The chemical reactions and pathways involving a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
cyclic nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
di-, tri-valent inorganic cation transport
The directed movement of inorganic cations with a valency of two or three into, out of, within or between cells by means of some external agent such as a transporter or pore. Inorganic cations are atoms or small molecules with a positive charge which do not contain carbon in covalent linkage.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
second-messenger-mediated signaling
A series of molecular signals in which an ion or small molecule is formed or released into the cytosol, thereby helping relay the signal within the cell.
cAMP-mediated signaling
A series of molecular signals in which a cell uses cyclic AMP to convert an extracellular signal into a response.
cyclic-nucleotide-mediated signaling
A series of molecular signals in which a cell uses a cyclic nucleotide to convert an extracellular signal into a response.
neurogenesis
Generation of cells within the nervous system.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
intracellular signaling pathway
The series of molecular events whereby information is sent from one location to another within a cell.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
cell projection organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
regulation of cyclic nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving cyclic nucleotides.
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
regulation of cAMP metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
neuron projection development
The process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites).
regulation of cyclase activity
Any process that modulates the frequency, rate or extent of cyclase activity.
negative regulation of cyclase activity
Any process that stops or reduces the activity of a cyclase.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
cell part morphogenesis
The process by which the anatomical structures of a cell part are generated and organized. Morphogenesis pertains to the creation of form.
nucleobase, nucleoside and nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleobases, nucleosides and nucleotides.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleobases, nucleosides, nucleotides and nucleic acids.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
negative regulation of catalytic activity
Any process that stops or reduces the activity of an enzyme.
negative regulation of molecular function
Any process that stops or reduces the rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular nitrogen compound biosynthetic process
The chemical reactions and pathways resulting in the formation of organic and inorganic nitrogenous compounds.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule biosynthetic process
The chemical reactions and pathways resulting in the formation of small molecules, any monomeric molecule of small relative molecular mass.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
heterocycle metabolic process
The chemical reactions and pathways involving heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
neuron development
The process whose specific outcome is the progression of a neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell.
cell morphogenesis involved in neuron differentiation
The process by which the structures of a neuron are generated and organized. This process occurs while the initially relatively unspecialized cell is acquiring the specialized features of a neuron.
generation of neurons
The process by which nerve cells are generated. This includes the production of neuroblasts and their differentiation into neurons.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
neuron projection morphogenesis
The process by which the anatomical structures of a neuron projection are generated and organized. Morphogenesis pertains to the creation of form. A neuron projection is any process extending from a neural cell, such as axons or dendrites.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cognition
The operation of the mind by which an organism becomes aware of objects of thought or perception; it includes the mental activities associated with thinking, learning, and memory.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of lyase activity
Any process that modulates the frequency, rate or extent of lyase activity, the catalysis of the cleavage of C-C, C-O, C-N and other bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond. They differ from other enzymes in that two substrates are involved in one reaction direction, but only one in the other direction. When acting on the single substrate, a molecule is eliminated and this generates either a new double bond or a new ring.
negative regulation of lyase activity
Any process that stops or reduces the rate of lyase activity, the catalysis of the cleavage of C-C, C-O, C-N and other bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
divalent metal ion transport
The directed movement of divalent metal cations, any metal ion with a +2 electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cell projection organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
cell recognition
The process by which a cell in a multicellular organism interprets its surroundings.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule biosynthetic process
The chemical reactions and pathways resulting in the formation of small molecules, any monomeric molecule of small relative molecular mass.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular nitrogen compound biosynthetic process
The chemical reactions and pathways resulting in the formation of organic and inorganic nitrogenous compounds.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
nucleobase, nucleoside and nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleobases, nucleosides and nucleotides.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
neuron projection morphogenesis
The process by which the anatomical structures of a neuron projection are generated and organized. Morphogenesis pertains to the creation of form. A neuron projection is any process extending from a neural cell, such as axons or dendrites.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
cell morphogenesis involved in differentiation
The change in form (cell shape and size) that occurs when relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
negative regulation of catalytic activity
Any process that stops or reduces the activity of an enzyme.
nucleobase, nucleoside, nucleotide and nucleic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleobases, nucleosides, nucleotides and nucleic acids.
negative regulation of cyclase activity
Any process that stops or reduces the activity of a cyclase.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
negative regulation of lyase activity
Any process that stops or reduces the rate of lyase activity, the catalysis of the cleavage of C-C, C-O, C-N and other bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond.
nucleobase, nucleoside and nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleobases, nucleosides and nucleotides.
neuron development
The process whose specific outcome is the progression of a neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell.
neuron recognition
The process by which a neuronal cell in a multicellular organism interprets its surroundings.
neuron projection development
The process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites).
cell morphogenesis involved in neuron differentiation
The process by which the structures of a neuron are generated and organized. This process occurs while the initially relatively unspecialized cell is acquiring the specialized features of a neuron.
learning or memory
The acquisition and processing of information and/or the storage and retrieval of this information over time.
neurogenesis
Generation of cells within the nervous system.
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
purine nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
negative regulation of adenylate cyclase activity
Any process that stops, prevents or reduces the frequency, rate or extent of adenylate cyclase activity.
negative regulation of adenylate cyclase activity
Any process that stops, prevents or reduces the frequency, rate or extent of adenylate cyclase activity.
cAMP biosynthetic process
The chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
purine nucleotide metabolic process
The chemical reactions and pathways involving a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleotides, any nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic-nucleotides (nucleoside cyclic phosphates).
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
axonogenesis
Generation of a long process of a neuron, that carries efferent (outgoing) action potentials from the cell body towards target cells.
divalent metal ion transport
The directed movement of divalent metal cations, any metal ion with a +2 electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
regulation of cAMP metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
nucleoside monophosphate biosynthetic process
The chemical reactions and pathways resulting in the formation of a nucleoside monophosphate, a glycosamine consisting of a base linked to a deoxyribose or ribose sugar esterified with phosphate on its glycose moiety.
G-protein signaling, coupled to cyclic nucleotide second messenger
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand, followed by modulation of a nucleotide cyclase activity and a subsequent change in the concentration of a cyclic nucleotide.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
cAMP biosynthetic process
The chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
cAMP metabolic process
The chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
cyclic nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
regulation of cyclic nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving cyclic nucleotides.
G-protein signaling, coupled to cAMP nucleotide second messenger
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand, followed by modulation of adenylyl cyclase activity and a subsequent change in the concentration of cyclic AMP.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endocytic vesicle
A membrane-bounded intracellular vesicle formed by invagination of the plasma membrane around an extracellular substance.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
synaptic vesicle
A secretory organelle, some 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and is secreted these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
postsynaptic density
The post synaptic density is a region that lies adjacent to the cytoplasmic face of the postsynaptic membrane at excitatory synapse. It forms a disc that consists of a range of proteins with different functions, some of which contact the cytoplasmic domains of ion channels in the postsynaptic membrane. The proteins making up the disc include receptors, and structural proteins linked to the actin cytoskeleton. They also include signalling machinery, such as protein kinases and phosphatases.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cell junction
A plasma membrane part that forms a specialized region of connection between two cells or between a cell and the extracellular matrix. At a cell junction, anchoring proteins extend through the plasma membrane to link cytoskeletal proteins in one cell to cytoskeletal proteins in neighboring cells or to proteins in the extracellular matrix.
coated vesicle
Small membrane-bounded organelle formed by pinching off of a coated region of membrane. Some coats are made of clathrin, whereas others are made from other proteins.
clathrin-coated vesicle
A vesicle with a coat formed of clathrin connected to the membrane via one of the clathrin adaptor complexes.
axon
The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter.
dendrite
A neuron projection that has a short, tapering, often branched, morphology, receives and integrates signals from other neurons or from sensory stimuli, and conducts a nerve impulse towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
coated vesicle membrane
The lipid bilayer surrounding a coated vesicle.
clathrin coated vesicle membrane
The lipid bilayer surrounding a clathrin-coated vesicle.
endocytic vesicle membrane
The lipid bilayer surrounding an endocytic vesicle.
synaptic vesicle membrane
The lipid bilayer surrounding a synaptic vesicle.
organelle membrane
The lipid bilayer surrounding an organelle.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
potassium channel complex
An ion channel complex through which potassium ions pass.
presynaptic membrane
A specialized area of membrane of the axon terminal that faces the plasma membrane of the neuron or muscle fiber with which the axon terminal establishes a synaptic junction; many synaptic junctions exhibit structural presynaptic characteristics, such as conical, electron-dense internal protrusions, that distinguish it from the remainder of the axon plasma membrane.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
neuron projection
A prolongation or process extending from a nerve cell, e.g. an axon or dendrite.
neuronal cell body
The portion of a neuron that includes the nucleus, but excludes all cell projections such as axons and dendrites.
dendritic spine
Protrusion from a dendrite. Spines are specialised subcellular compartments involved in the synaptic transmission. They are linked to the dendritic shaft by a restriction. Because of their bulb shape, they function as a biochemical and an electrical compartment. Spine remodeling is though to be involved in synaptic plasticity.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
cell body
The portion of a cell bearing surface projections such as axons, dendrites, cilia, or flagella that includes the nucleus, but excludes all cell projections.
neuron spine
A small membranous protrusion, often ending in a bulbous head and attached to the neuron by a narrow stalk or neck.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse
The junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell; the site of interneuronal communication. As the nerve fiber approaches the synapse it enlarges into a specialized structure, the presynaptic nerve ending, which contains mitochondria and synaptic vesicles. At the tip of the nerve ending is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic nerve ending secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane.
postsynaptic membrane
A specialized area of membrane facing the presynaptic membrane on the tip of the nerve ending and separated from it by a minute cleft (the synaptic cleft). Neurotransmitters across the synaptic cleft and transmit the signal to the postsynaptic membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
organelle membrane
The lipid bilayer surrounding an organelle.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
presynaptic membrane
A specialized area of membrane of the axon terminal that faces the plasma membrane of the neuron or muscle fiber with which the axon terminal establishes a synaptic junction; many synaptic junctions exhibit structural presynaptic characteristics, such as conical, electron-dense internal protrusions, that distinguish it from the remainder of the axon plasma membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
postsynaptic membrane
A specialized area of membrane facing the presynaptic membrane on the tip of the nerve ending and separated from it by a minute cleft (the synaptic cleft). Neurotransmitters across the synaptic cleft and transmit the signal to the postsynaptic membrane.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
postsynaptic density
The post synaptic density is a region that lies adjacent to the cytoplasmic face of the postsynaptic membrane at excitatory synapse. It forms a disc that consists of a range of proteins with different functions, some of which contact the cytoplasmic domains of ion channels in the postsynaptic membrane. The proteins making up the disc include receptors, and structural proteins linked to the actin cytoskeleton. They also include signalling machinery, such as protein kinases and phosphatases.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
dendritic spine
Protrusion from a dendrite. Spines are specialised subcellular compartments involved in the synaptic transmission. They are linked to the dendritic shaft by a restriction. Because of their bulb shape, they function as a biochemical and an electrical compartment. Spine remodeling is though to be involved in synaptic plasticity.
dendritic spine
Protrusion from a dendrite. Spines are specialised subcellular compartments involved in the synaptic transmission. They are linked to the dendritic shaft by a restriction. Because of their bulb shape, they function as a biochemical and an electrical compartment. Spine remodeling is though to be involved in synaptic plasticity.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
endocytic vesicle membrane
The lipid bilayer surrounding an endocytic vesicle.
coated vesicle membrane
The lipid bilayer surrounding a coated vesicle.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
synaptic vesicle
A secretory organelle, some 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and is secreted these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane.
clathrin coated vesicle membrane
The lipid bilayer surrounding a clathrin-coated vesicle.
synaptic vesicle membrane
The lipid bilayer surrounding a synaptic vesicle.

protein binding
Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
peptide receptor activity
Combining with an extracellular or intracellular peptide to initiate a change in cell activity.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively and non-covalently with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
hormone activity
The action characteristic of a hormone, any substance formed in very small amounts in one specialized organ or group of cells and carried (sometimes in the bloodstream) to another organ or group of cells in the same organism, upon which it has a specific regulatory action. The term was originally applied to agents with a stimulatory physiological action in vertebrate animals (as opposed to a chalone, which has a depressant action). Usage is now extended to regulatory compounds in lower animals and plants, and to synthetic substances having comparable effects; all bind receptors and trigger some biological process.
neuropeptide hormone activity
The action characteristic of a neuropeptide hormone, any peptide hormone that acts in the central nervous system. A neuropeptide is any of several types of molecules found in brain tissue, composed of short chains of amino acids; they include endorphins, enkephalins, vasopressin, and others. They are often localized in axon terminals at synapses and are classified as putative neurotransmitters, although some are also hormones.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
extracellular ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific extracellular ligand has been bound by the channel complex or one of its constituent parts.
voltage-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons.
voltage-gated potassium channel activity
Catalysis of the transmembrane transfer of a potassium ion by a voltage-gated channel.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
potassium channel activity
Catalysis of facilitated diffusion of a potassium ion (by an energy-independent process) involving passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
glutamate receptor activity
Combining with glutamate to initiate a change in cell activity.
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
peptide receptor activity, G-protein coupled
Combining with an extracellular or intracellular peptide to initiate a G-protein mediated change in cell activity. A G-protein is a signal transduction molecule that alternates between an inactive GDP-bound and an active GTP-bound state.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
voltage-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel whose open state is dependent on the voltage across the membrane in which it is embedded.
ligand-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
voltage-gated cation channel activity
Catalysis of the transmembrane transfer of a cation by a voltage-gated channel. A cation is a positively charged ion.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
neurotransmitter binding
Interacting selectively and non-covalently with a neurotransmitter, any chemical substance that is capable of transmitting (or inhibiting the transmission of) a nerve impulse from a neuron to another cell.
peptide binding
Interacting selectively and non-covalently with peptides, any of a group of organic compounds comprising two or more amino acids linked by peptide bonds.
neuropeptide binding
Interacting selectively and non-covalently and stoichiometrically with neuropeptides, peptides with direct synaptic effects (peptide neurotransmitters) or indirect modulatory effects on the nervous system (peptide neuromodulators).
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
peptide receptor activity
Combining with an extracellular or intracellular peptide to initiate a change in cell activity.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
peptide receptor activity, G-protein coupled
Combining with an extracellular or intracellular peptide to initiate a G-protein mediated change in cell activity. A G-protein is a signal transduction molecule that alternates between an inactive GDP-bound and an active GTP-bound state.
voltage-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
voltage-gated cation channel activity
Catalysis of the transmembrane transfer of a cation by a voltage-gated channel. A cation is a positively charged ion.
voltage-gated potassium channel activity
Catalysis of the transmembrane transfer of a potassium ion by a voltage-gated channel.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 2.477e-07 | 6.46 | 25 | 102 | Neuroactive ligand-receptor interaction |
| 04020 | 1.069e-03 | 4.306 | 15 | 68 | Calcium signaling pathway |
| 04720 | 5.946e-03 | 1.583 | 8 | 25 | Long-term potentiation |
ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (ENSG00000172350), score: 0.48 ACCN4amiloride-sensitive cation channel 4, pituitary (ENSG00000072182), score: 0.6 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.5 ADAM22ADAM metallopeptidase domain 22 (ENSG00000008277), score: 0.48 ADCY8adenylate cyclase 8 (brain) (ENSG00000155897), score: 0.52 ADCYAP1adenylate cyclase activating polypeptide 1 (pituitary) (ENSG00000141433), score: 0.49 AK2adenylate kinase 2 (ENSG00000004455), score: -0.45 ALKanaplastic lymphoma receptor tyrosine kinase (ENSG00000171094), score: 0.52 AMZ1archaelysin family metallopeptidase 1 (ENSG00000174945), score: 0.48 ANKFN1ankyrin-repeat and fibronectin type III domain containing 1 (ENSG00000153930), score: 0.53 ANKRA2ankyrin repeat, family A (RFXANK-like), 2 (ENSG00000164331), score: 0.56 ANKRD46ankyrin repeat domain 46 (ENSG00000186106), score: 0.51 ANO3anoctamin 3 (ENSG00000134343), score: 0.66 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: -0.52 ARCactivity-regulated cytoskeleton-associated protein (ENSG00000198576), score: 0.54 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (ENSG00000149182), score: -0.46 ARHGAP32Rho GTPase activating protein 32 (ENSG00000134909), score: 0.52 ARPP21cAMP-regulated phosphoprotein, 21kDa (ENSG00000172995), score: 0.56 ASTN1astrotactin 1 (ENSG00000152092), score: 0.49 ATP13A5ATPase type 13A5 (ENSG00000187527), score: 0.58 ATP2B1ATPase, Ca++ transporting, plasma membrane 1 (ENSG00000070961), score: 0.56 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (ENSG00000113732), score: -0.42 AVPR1Barginine vasopressin receptor 1B (ENSG00000198049), score: 0.47 B3GALT1UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 (ENSG00000172318), score: 0.61 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.68 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (ENSG00000086062), score: -0.42 B4GALT6UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (ENSG00000118276), score: 0.48 BAI1brain-specific angiogenesis inhibitor 1 (ENSG00000181790), score: 0.5 BCAT1branched chain amino-acid transaminase 1, cytosolic (ENSG00000060982), score: 0.48 BCL11BB-cell CLL/lymphoma 11B (zinc finger protein) (ENSG00000127152), score: 0.82 BMP3bone morphogenetic protein 3 (ENSG00000152785), score: 0.75 BRS3bombesin-like receptor 3 (ENSG00000102239), score: 0.75 BTBD8BTB (POZ) domain containing 8 (ENSG00000189195), score: 0.62 C13orf36chromosome 13 open reading frame 36 (ENSG00000180440), score: 0.48 C14orf159chromosome 14 open reading frame 159 (ENSG00000133943), score: -0.43 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.55 C1orf226chromosome 1 open reading frame 226 (ENSG00000198929), score: 0.47 C1orf58chromosome 1 open reading frame 58 (ENSG00000162819), score: 0.49 C20orf103chromosome 20 open reading frame 103 (ENSG00000125869), score: 0.58 C20orf186chromosome 20 open reading frame 186 (ENSG00000186191), score: 0.48 C22orf13chromosome 22 open reading frame 13 (ENSG00000138867), score: -0.42 C8orf34chromosome 8 open reading frame 34 (ENSG00000165084), score: 0.76 C9orf4chromosome 9 open reading frame 4 (ENSG00000136805), score: 0.49 CACNA1Ecalcium channel, voltage-dependent, R type, alpha 1E subunit (ENSG00000198216), score: 0.59 CACNB4calcium channel, voltage-dependent, beta 4 subunit (ENSG00000182389), score: 0.48 CACNG3calcium channel, voltage-dependent, gamma subunit 3 (ENSG00000006116), score: 0.6 CACNG4calcium channel, voltage-dependent, gamma subunit 4 (ENSG00000075461), score: 0.5 CADM2cell adhesion molecule 2 (ENSG00000175161), score: 0.48 CAMK2Acalcium/calmodulin-dependent protein kinase II alpha (ENSG00000070808), score: 0.52 CASD1CAS1 domain containing 1 (ENSG00000127995), score: 0.49 CASP6caspase 6, apoptosis-related cysteine peptidase (ENSG00000138794), score: -0.42 CASTcalpastatin (ENSG00000153113), score: -0.49 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: -0.44 CD44CD44 molecule (Indian blood group) (ENSG00000026508), score: -0.44 CDH10cadherin 10, type 2 (T2-cadherin) (ENSG00000040731), score: 0.5 CDH12cadherin 12, type 2 (N-cadherin 2) (ENSG00000154162), score: 0.49 CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.52 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.49 CDH9cadherin 9, type 2 (T1-cadherin) (ENSG00000113100), score: 0.57 CDK17cyclin-dependent kinase 17 (ENSG00000059758), score: 0.64 CDKL5cyclin-dependent kinase-like 5 (ENSG00000008086), score: 0.53 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.59 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: -0.55 CHRM3cholinergic receptor, muscarinic 3 (ENSG00000133019), score: 0.49 CLCN3chloride channel 3 (ENSG00000109572), score: 0.54 CLVS1clavesin 1 (ENSG00000177182), score: 0.49 CLVS2clavesin 2 (ENSG00000146352), score: 0.51 CNKSR2connector enhancer of kinase suppressor of Ras 2 (ENSG00000149970), score: 0.52 CNTN3contactin 3 (plasmacytoma associated) (ENSG00000113805), score: 0.68 CNTN4contactin 4 (ENSG00000144619), score: 0.52 CNTN5contactin 5 (ENSG00000149972), score: 0.72 COL19A1collagen, type XIX, alpha 1 (ENSG00000082293), score: 0.55 COL25A1collagen, type XXV, alpha 1 (ENSG00000188517), score: 0.75 CPNE4copine IV (ENSG00000196353), score: 0.56 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.46 CRHcorticotropin releasing hormone (ENSG00000147571), score: 0.5 CSMD2CUB and Sushi multiple domains 2 (ENSG00000121904), score: 0.63 CSRNP3cysteine-serine-rich nuclear protein 3 (ENSG00000178662), score: 0.49 CTNND1catenin (cadherin-associated protein), delta 1 (ENSG00000198561), score: -0.58 CTTNBP2cortactin binding protein 2 (ENSG00000077063), score: 0.56 CUL4Acullin 4A (ENSG00000139842), score: -0.43 D2HGDHD-2-hydroxyglutarate dehydrogenase (ENSG00000180902), score: -0.42 DAZAP1DAZ associated protein 1 (ENSG00000071626), score: -0.43 DBC1deleted in bladder cancer 1 (ENSG00000078725), score: 0.48 DCLK1doublecortin-like kinase 1 (ENSG00000133083), score: 0.48 DCLK3doublecortin-like kinase 3 (ENSG00000163673), score: 0.68 DCNdecorin (ENSG00000011465), score: -0.43 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.7 DLGAP2discs, large (Drosophila) homolog-associated protein 2 (ENSG00000198010), score: 0.62 DLK1delta-like 1 homolog (Drosophila) (ENSG00000185559), score: 0.49 DLX1distal-less homeobox 1 (ENSG00000144355), score: 0.53 DLX6distal-less homeobox 6 (ENSG00000006377), score: 0.74 DNAJC27DnaJ (Hsp40) homolog, subfamily C, member 27 (ENSG00000115137), score: 0.48 DRD2dopamine receptor D2 (ENSG00000149295), score: 0.64 DRP2dystrophin related protein 2 (ENSG00000102385), score: 0.53 DSCAMDown syndrome cell adhesion molecule (ENSG00000171587), score: 0.58 DSCAML1Down syndrome cell adhesion molecule like 1 (ENSG00000177103), score: 0.49 DUSP19dual specificity phosphatase 19 (ENSG00000162999), score: 0.56 EFR3BEFR3 homolog B (S. cerevisiae) (ENSG00000084710), score: 0.47 EIF2B4eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa (ENSG00000115211), score: -0.43 ELFN2extracellular leucine-rich repeat and fibronectin type III domain containing 2 (ENSG00000166897), score: 0.49 ELMOD1ELMO/CED-12 domain containing 1 (ENSG00000110675), score: 0.52 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (ENSG00000118402), score: 0.47 EPHA3EPH receptor A3 (ENSG00000044524), score: 0.54 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.75 EPHA7EPH receptor A7 (ENSG00000135333), score: 0.61 EPHX4epoxide hydrolase 4 (ENSG00000172031), score: 0.66 ERBB2v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) (ENSG00000141736), score: -0.42 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (ENSG00000178568), score: 0.47 FADDFas (TNFRSF6)-associated via death domain (ENSG00000168040), score: -0.53 FAM105Afamily with sequence similarity 105, member A (ENSG00000145569), score: 0.57 FAM164Afamily with sequence similarity 164, member A (ENSG00000104427), score: 0.52 FAM19A1family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 (ENSG00000183662), score: 0.55 FAM49Afamily with sequence similarity 49, member A (ENSG00000197872), score: 0.48 FAM5Bfamily with sequence similarity 5, member B (ENSG00000198797), score: 0.51 FAM5Cfamily with sequence similarity 5, member C (ENSG00000162670), score: 0.56 FAM65Bfamily with sequence similarity 65, member B (ENSG00000111913), score: 0.49 FEZF2FEZ family zinc finger 2 (ENSG00000153266), score: 0.53 FIBCD1fibrinogen C domain containing 1 (ENSG00000130720), score: 0.81 FOXG1forkhead box G1 (ENSG00000176165), score: 0.47 FOXP2forkhead box P2 (ENSG00000128573), score: 0.48 FRMPD4FERM and PDZ domain containing 4 (ENSG00000169933), score: 0.56 GABRA1gamma-aminobutyric acid (GABA) A receptor, alpha 1 (ENSG00000022355), score: 0.47 GABRA2gamma-aminobutyric acid (GABA) A receptor, alpha 2 (ENSG00000151834), score: 0.66 GABRA4gamma-aminobutyric acid (GABA) A receptor, alpha 4 (ENSG00000109158), score: 0.64 GAD2glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) (ENSG00000136750), score: 0.62 GALNTL6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 (ENSG00000174473), score: 0.72 GALR1galanin receptor 1 (ENSG00000166573), score: 0.63 GBX2gastrulation brain homeobox 2 (ENSG00000168505), score: 0.56 GDPD1glycerophosphodiester phosphodiesterase domain containing 1 (ENSG00000153982), score: 0.47 GLRA2glycine receptor, alpha 2 (ENSG00000101958), score: 0.56 GLRA3glycine receptor, alpha 3 (ENSG00000145451), score: 0.59 GNALguanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type (ENSG00000141404), score: 0.53 GPM6Bglycoprotein M6B (ENSG00000046653), score: 0.48 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 0.91 GPRC5CG protein-coupled receptor, family C, group 5, member C (ENSG00000170412), score: -0.46 GRIA1glutamate receptor, ionotropic, AMPA 1 (ENSG00000155511), score: 0.51 GRIA2glutamate receptor, ionotropic, AMPA 2 (ENSG00000120251), score: 0.49 GRIA3glutamate receptor, ionotrophic, AMPA 3 (ENSG00000125675), score: 0.47 GRIN2Bglutamate receptor, ionotropic, N-methyl D-aspartate 2B (ENSG00000150086), score: 0.75 GRIN3Aglutamate receptor, ionotropic, N-methyl-D-aspartate 3A (ENSG00000198785), score: 0.58 GRIP1glutamate receptor interacting protein 1 (ENSG00000155974), score: 0.51 GRM3glutamate receptor, metabotropic 3 (ENSG00000198822), score: 0.5 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: -0.44 HEATR5BHEAT repeat containing 5B (ENSG00000008869), score: 0.48 HMP19HMP19 protein (ENSG00000170091), score: 0.48 HS2ST1heparan sulfate 2-O-sulfotransferase 1 (ENSG00000153936), score: 0.51 HTR1D5-hydroxytryptamine (serotonin) receptor 1D (ENSG00000179546), score: 0.71 HTR45-hydroxytryptamine (serotonin) receptor 4 (ENSG00000164270), score: 0.79 IAH1isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) (ENSG00000134330), score: -0.47 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.69 IL1RAPL2interleukin 1 receptor accessory protein-like 2 (ENSG00000189108), score: 0.87 ILDR2immunoglobulin-like domain containing receptor 2 (ENSG00000143195), score: 0.55 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: 0.59 KCNA3potassium voltage-gated channel, shaker-related subfamily, member 3 (ENSG00000177272), score: 0.49 KCNA4potassium voltage-gated channel, shaker-related subfamily, member 4 (ENSG00000182255), score: 0.68 KCNA6potassium voltage-gated channel, shaker-related subfamily, member 6 (ENSG00000151079), score: 0.47 KCNG3potassium voltage-gated channel, subfamily G, member 3 (ENSG00000171126), score: 0.6 KCNH7potassium voltage-gated channel, subfamily H (eag-related), member 7 (ENSG00000184611), score: 0.68 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.59 KCNK2potassium channel, subfamily K, member 2 (ENSG00000082482), score: 0.76 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (ENSG00000184156), score: 0.63 KCNS2potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 (ENSG00000156486), score: 0.72 KCTD16potassium channel tetramerisation domain containing 16 (ENSG00000183775), score: 0.51 KCTD4potassium channel tetramerisation domain containing 4 (ENSG00000180332), score: 0.67 KIAA0391KIAA0391 (ENSG00000100890), score: -0.45 KIAA1279KIAA1279 (ENSG00000198954), score: 0.5 KIAA1467KIAA1467 (ENSG00000084444), score: 0.53 KIAA2022KIAA2022 (ENSG00000050030), score: 0.65 KRT9keratin 9 (ENSG00000171403), score: 0.66 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.55 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.58 LMO1LIM domain only 1 (rhombotin 1) (ENSG00000166407), score: 0.61 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.59 LOC100293817similar to hCG1811893 (ENSG00000144460), score: 0.5 LPPR4lipid phosphate phosphatase-related protein type 4 (ENSG00000117600), score: 0.59 LRFN2leucine rich repeat and fibronectin type III domain containing 2 (ENSG00000156564), score: 0.47 LRFN5leucine rich repeat and fibronectin type III domain containing 5 (ENSG00000165379), score: 0.59 LRP12low density lipoprotein receptor-related protein 12 (ENSG00000147650), score: 0.49 LRRC40leucine rich repeat containing 40 (ENSG00000066557), score: 0.64 LRRC4Cleucine rich repeat containing 4C (ENSG00000148948), score: 0.55 LRRC7leucine rich repeat containing 7 (ENSG00000033122), score: 0.79 LRRTM3leucine rich repeat transmembrane neuronal 3 (ENSG00000198739), score: 0.48 LRTM2leucine-rich repeats and transmembrane domains 2 (ENSG00000166159), score: 0.49 LSM10LSM10, U7 small nuclear RNA associated (ENSG00000181817), score: -0.44 MAML1mastermind-like 1 (Drosophila) (ENSG00000161021), score: -0.49 MAP2K1mitogen-activated protein kinase kinase 1 (ENSG00000169032), score: 0.54 MAP3K3mitogen-activated protein kinase kinase kinase 3 (ENSG00000198909), score: -0.46 MAP7D2MAP7 domain containing 2 (ENSG00000184368), score: 0.48 MAPK8mitogen-activated protein kinase 8 (ENSG00000107643), score: 0.52 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: -0.43 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (ENSG00000162889), score: -0.45 MAS1MAS1 oncogene (ENSG00000130368), score: 0.52 MC4Rmelanocortin 4 receptor (ENSG00000166603), score: 0.57 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.65 MEF2Cmyocyte enhancer factor 2C (ENSG00000081189), score: 0.48 MKL2MKL/myocardin-like 2 (ENSG00000186260), score: 0.57 MLXIPMLX interacting protein (ENSG00000175727), score: -0.55 MRPL21mitochondrial ribosomal protein L21 (ENSG00000197345), score: -0.43 MYH9myosin, heavy chain 9, non-muscle (ENSG00000100345), score: -0.42 MYO16myosin XVI (ENSG00000041515), score: 0.58 NCKAP1NCK-associated protein 1 (ENSG00000061676), score: 0.56 NDST4N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 (ENSG00000138653), score: 0.73 NECAB1N-terminal EF-hand calcium binding protein 1 (ENSG00000123119), score: 0.56 NEK9NIMA (never in mitosis gene a)- related kinase 9 (ENSG00000119638), score: -0.47 NELL2NEL-like 2 (chicken) (ENSG00000184613), score: 0.47 NETO1neuropilin (NRP) and tolloid (TLL)-like 1 (ENSG00000166342), score: 0.61 NFE2L2nuclear factor (erythroid-derived 2)-like 2 (ENSG00000116044), score: -0.43 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (ENSG00000109320), score: -0.47 NLGN1neuroligin 1 (ENSG00000169760), score: 0.57 NOL4nucleolar protein 4 (ENSG00000101746), score: 0.48 NOVnephroblastoma overexpressed gene (ENSG00000136999), score: 0.48 NPY2Rneuropeptide Y receptor Y2 (ENSG00000185149), score: 1 NPY5Rneuropeptide Y receptor Y5 (ENSG00000164129), score: 0.58 NRIP3nuclear receptor interacting protein 3 (ENSG00000175352), score: 0.47 NRSN1neurensin 1 (ENSG00000152954), score: 0.52 NTNG1netrin G1 (ENSG00000162631), score: 0.48 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (ENSG00000148053), score: 0.47 NTSneurotensin (ENSG00000133636), score: 0.93 NTSR1neurotensin receptor 1 (high affinity) (ENSG00000101188), score: 0.56 OLIG3oligodendrocyte transcription factor 3 (ENSG00000177468), score: 0.72 OPCMLopioid binding protein/cell adhesion molecule-like (ENSG00000183715), score: 0.52 OPRK1opioid receptor, kappa 1 (ENSG00000082556), score: 0.7 OSTF1osteoclast stimulating factor 1 (ENSG00000134996), score: -0.55 OTOFotoferlin (ENSG00000115155), score: 0.7 P2RY12purinergic receptor P2Y, G-protein coupled, 12 (ENSG00000169313), score: 0.56 PCDH19protocadherin 19 (ENSG00000165194), score: 0.55 PCDH20protocadherin 20 (ENSG00000197991), score: 0.65 PCSK1proprotein convertase subtilisin/kexin type 1 (ENSG00000175426), score: 0.48 PCSK2proprotein convertase subtilisin/kexin type 2 (ENSG00000125851), score: 0.52 PDE10Aphosphodiesterase 10A (ENSG00000112541), score: 0.6 PDE7Bphosphodiesterase 7B (ENSG00000171408), score: 0.53 PDS5BPDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) (ENSG00000083642), score: 0.5 PENKproenkephalin (ENSG00000181195), score: 0.6 PEX5Lperoxisomal biogenesis factor 5-like (ENSG00000114757), score: 0.56 PIK3C3phosphoinositide-3-kinase, class 3 (ENSG00000078142), score: 0.5 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (ENSG00000182621), score: 0.63 PNOCprepronociceptin (ENSG00000168081), score: 0.48 POU1F1POU class 1 homeobox 1 (ENSG00000064835), score: 0.59 PPM1Eprotein phosphatase, Mg2+/Mn2+ dependent, 1E (ENSG00000175175), score: 0.52 PPP3CAprotein phosphatase 3, catalytic subunit, alpha isozyme (ENSG00000138814), score: 0.56 PQLC1PQ loop repeat containing 1 (ENSG00000122490), score: -0.41 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.45 PRICKLE1prickle homolog 1 (Drosophila) (ENSG00000139174), score: 0.49 PRICKLE2prickle homolog 2 (Drosophila) (ENSG00000163637), score: 0.54 PRKG2protein kinase, cGMP-dependent, type II (ENSG00000138669), score: 0.57 PSKH1protein serine kinase H1 (ENSG00000159792), score: -0.46 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: -0.49 R3HDM1R3H domain containing 1 (ENSG00000048991), score: 0.54 RAB3GAP2RAB3 GTPase activating protein subunit 2 (non-catalytic) (ENSG00000118873), score: 0.58 RALGAPA1Ral GTPase activating protein, alpha subunit 1 (catalytic) (ENSG00000174373), score: 0.51 RASD2RASD family, member 2 (ENSG00000100302), score: 0.53 RASGEF1ARasGEF domain family, member 1A (ENSG00000198915), score: 0.53 RGS20regulator of G-protein signaling 20 (ENSG00000147509), score: 0.56 RGS7BPregulator of G-protein signaling 7 binding protein (ENSG00000186479), score: 0.54 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.56 RGS9regulator of G-protein signaling 9 (ENSG00000108370), score: 0.6 RPE65retinal pigment epithelium-specific protein 65kDa (ENSG00000116745), score: 0.48 RPS6KC1ribosomal protein S6 kinase, 52kDa, polypeptide 1 (ENSG00000136643), score: 0.49 RSPO3R-spondin 3 homolog (Xenopus laevis) (ENSG00000146374), score: 0.5 RXFP3relaxin/insulin-like family peptide receptor 3 (ENSG00000182631), score: 0.53 SCAMP1secretory carrier membrane protein 1 (ENSG00000085365), score: 0.62 SCG2secretogranin II (ENSG00000171951), score: 0.58 SEMA3Asema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A (ENSG00000075213), score: 0.76 SEMA3Esema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E (ENSG00000170381), score: 0.52 SERTAD4SERTA domain containing 4 (ENSG00000082497), score: 0.5 SGCZsarcoglycan, zeta (ENSG00000185053), score: 0.48 SGTBsmall glutamine-rich tetratricopeptide repeat (TPR)-containing, beta (ENSG00000197860), score: 0.52 SLC1A2solute carrier family 1 (glial high affinity glutamate transporter), member 2 (ENSG00000110436), score: 0.47 SLC24A4solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 (ENSG00000140090), score: 0.6 SLC25A37solute carrier family 25, member 37 (ENSG00000147454), score: -0.51 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.56 SLC39A13solute carrier family 39 (zinc transporter), member 13 (ENSG00000165915), score: -0.49 SLC41A3solute carrier family 41, member 3 (ENSG00000114544), score: -0.42 SLC4A10solute carrier family 4, sodium bicarbonate transporter, member 10 (ENSG00000144290), score: 0.52 SLC5A7solute carrier family 5 (choline transporter), member 7 (ENSG00000115665), score: 0.95 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (ENSG00000132164), score: 0.55 SLC7A14solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 (ENSG00000013293), score: 0.54 SNAP23synaptosomal-associated protein, 23kDa (ENSG00000092531), score: -0.46 SNTG1syntrophin, gamma 1 (ENSG00000147481), score: 0.57 SPRED1sprouty-related, EVH1 domain containing 1 (ENSG00000166068), score: 0.59 SSTsomatostatin (ENSG00000157005), score: 0.64 ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (ENSG00000117069), score: 0.53 STIM2stromal interaction molecule 2 (ENSG00000109689), score: 0.52 STK40serine/threonine kinase 40 (ENSG00000196182), score: -0.42 STX12syntaxin 12 (ENSG00000117758), score: 0.5 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.5 SV2Csynaptic vesicle glycoprotein 2C (ENSG00000122012), score: 0.78 SYNJ1synaptojanin 1 (ENSG00000159082), score: 0.61 SYT11synaptotagmin XI (ENSG00000132718), score: 0.47 TAC1tachykinin, precursor 1 (ENSG00000006128), score: 0.83 TBC1D19TBC1 domain family, member 19 (ENSG00000109680), score: 0.49 TBC1D30TBC1 domain family, member 30 (ENSG00000111490), score: 0.58 TFAP2Dtranscription factor AP-2 delta (activating enhancer binding protein 2 delta) (ENSG00000008197), score: 0.8 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (ENSG00000198959), score: -0.43 THSD7Athrombospondin, type I, domain containing 7A (ENSG00000005108), score: 0.63 THY1Thy-1 cell surface antigen (ENSG00000154096), score: 0.49 TLN1talin 1 (ENSG00000137076), score: -0.47 TMEFF2transmembrane protein with EGF-like and two follistatin-like domains 2 (ENSG00000144339), score: 0.54 TMEM200Atransmembrane protein 200A (ENSG00000164484), score: 0.69 TMEM74transmembrane protein 74 (ENSG00000164841), score: 0.48 TOB2transducer of ERBB2, 2 (ENSG00000183864), score: -0.49 TPBGtrophoblast glycoprotein (ENSG00000146242), score: 0.55 TRABDTraB domain containing (ENSG00000170638), score: -0.45 TRAM2translocation associated membrane protein 2 (ENSG00000065308), score: -0.46 TRHRthyrotropin-releasing hormone receptor (ENSG00000174417), score: 0.71 TRPC1transient receptor potential cation channel, subfamily C, member 1 (ENSG00000144935), score: 0.56 TRPC5transient receptor potential cation channel, subfamily C, member 5 (ENSG00000072315), score: 0.81 TRPC6transient receptor potential cation channel, subfamily C, member 6 (ENSG00000137672), score: 0.59 TSPAN2tetraspanin 2 (ENSG00000134198), score: 0.54 UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.44 UNC5Dunc-5 homolog D (C. elegans) (ENSG00000156687), score: 0.53 USP14ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) (ENSG00000101557), score: 0.5 USP45ubiquitin specific peptidase 45 (ENSG00000123552), score: 0.51 VIPvasoactive intestinal peptide (ENSG00000146469), score: 0.56 VSTM2AV-set and transmembrane domain containing 2A (ENSG00000170419), score: 0.57 VWC2Lvon Willebrand factor C domain-containing protein 2-like (ENSG00000174453), score: 0.57 WDR7WD repeat domain 7 (ENSG00000091157), score: 0.48 WNT4wingless-type MMTV integration site family, member 4 (ENSG00000162552), score: 0.71 ZC3H12Bzinc finger CCCH-type containing 12B (ENSG00000102053), score: 0.52 ZNF395zinc finger protein 395 (ENSG00000186918), score: -0.46
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_br_f_ca1 | mdo | br | f | _ |
| mmu_br_m2_ca1 | mmu | br | m | 2 |
| oan_br_f_ca1 | oan | br | f | _ |
| mmu_br_f_ca1 | mmu | br | f | _ |
| mmu_br_m1_ca1 | mmu | br | m | 1 |
| oan_br_m_ca1 | oan | br | m | _ |
| gga_br_m_ca1 | gga | br | m | _ |
| gga_br_f_ca1 | gga | br | f | _ |