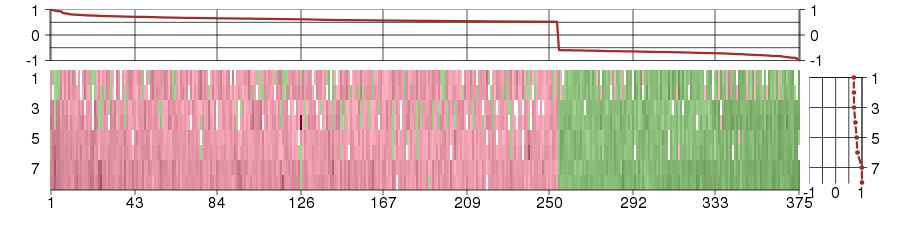

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
peroxisome
A small, membrane-bounded organelle that uses dioxygen (O2) to oxidize organic molecules; contains some enzymes that produce and others that degrade hydrogen peroxide (H2O2).
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
organelle membrane
The lipid bilayer surrounding an organelle.
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
carboxylic acid binding
Interacting selectively and non-covalently with a carboxylic acid, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
cofactor binding
Interacting selectively and non-covalently with a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 01100 | 6.272e-03 | 27.04 | 45 | 353 | Metabolic pathways |
| 00480 | 2.118e-02 | 0.766 | 5 | 10 | Glutathione metabolism |
| 04146 | 4.580e-02 | 2.298 | 8 | 30 | Peroxisome |
AARS2alanyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000124608), score: 0.64 ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (ENSG00000165029), score: 0.62 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (ENSG00000154265), score: -0.82 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.54 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: 0.52 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (ENSG00000160179), score: -0.65 ABHD12abhydrolase domain containing 12 (ENSG00000100997), score: -0.59 ABHD6abhydrolase domain containing 6 (ENSG00000163686), score: -0.73 ACAD8acyl-CoA dehydrogenase family, member 8 (ENSG00000151498), score: 0.57 ACBD5acyl-CoA binding domain containing 5 (ENSG00000107897), score: 0.54 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.71 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.82 ACO1aconitase 1, soluble (ENSG00000122729), score: 0.52 ACSL1acyl-CoA synthetase long-chain family member 1 (ENSG00000151726), score: 0.56 ACTL6Aactin-like 6A (ENSG00000136518), score: 0.67 ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.78 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.89 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.58 AHRaryl hydrocarbon receptor (ENSG00000106546), score: 0.57 AHSA2AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) (ENSG00000173209), score: -0.61 AICDAactivation-induced cytidine deaminase (ENSG00000111732), score: 0.62 AIFM2apoptosis-inducing factor, mitochondrion-associated, 2 (ENSG00000042286), score: 0.54 AK2adenylate kinase 2 (ENSG00000004455), score: 0.72 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.55 ALAS1aminolevulinate, delta-, synthase 1 (ENSG00000023330), score: 0.55 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (ENSG00000119711), score: 0.72 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.99 ANAPC2anaphase promoting complex subunit 2 (ENSG00000176248), score: -0.67 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.87 ANKRD22ankyrin repeat domain 22 (ENSG00000152766), score: 0.55 AP2B1adaptor-related protein complex 2, beta 1 subunit (ENSG00000006125), score: -0.69 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: 0.79 ARHGAP40Rho GTPase activating protein 40 (ENSG00000124143), score: 0.54 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 0.95 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.75 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.71 ASXL2additional sex combs like 2 (Drosophila) (ENSG00000143970), score: 0.65 ATG16L1ATG16 autophagy related 16-like 1 (S. cerevisiae) (ENSG00000085978), score: 0.63 ATG4AATG4 autophagy related 4 homolog A (S. cerevisiae) (ENSG00000101844), score: 0.67 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: 0.66 ATMINATM interactor (ENSG00000166454), score: -0.82 ATP13A2ATPase type 13A2 (ENSG00000159363), score: -0.66 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.81 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.82 ATXN2ataxin 2 (ENSG00000204842), score: 0.65 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.77 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (ENSG00000083123), score: 0.65 BMP2bone morphogenetic protein 2 (ENSG00000125845), score: 0.58 BRP44brain protein 44 (ENSG00000143158), score: 0.55 BTKBruton agammaglobulinemia tyrosine kinase (ENSG00000010671), score: 0.61 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.72 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.67 C14orf45chromosome 14 open reading frame 45 (ENSG00000119636), score: -0.64 C15orf59chromosome 15 open reading frame 59 (ENSG00000205363), score: -0.6 C1orf123chromosome 1 open reading frame 123 (ENSG00000162384), score: 0.65 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.87 C22orf13chromosome 22 open reading frame 13 (ENSG00000138867), score: 0.59 C3orf23chromosome 3 open reading frame 23 (ENSG00000179152), score: 0.66 C6orf150chromosome 6 open reading frame 150 (ENSG00000164430), score: 0.57 C6orf57chromosome 6 open reading frame 57 (ENSG00000154079), score: 0.54 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (ENSG00000163050), score: 0.65 CADM4cell adhesion molecule 4 (ENSG00000105767), score: -0.63 CALCRLcalcitonin receptor-like (ENSG00000064989), score: 0.53 CAMKK2calcium/calmodulin-dependent protein kinase kinase 2, beta (ENSG00000110931), score: -0.6 CAMSAP1calmodulin regulated spectrin-associated protein 1 (ENSG00000130559), score: -0.66 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.8 CBY1chibby homolog 1 (Drosophila) (ENSG00000100211), score: -0.69 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: 0.54 CCDC28Acoiled-coil domain containing 28A (ENSG00000024862), score: -0.66 CCDC82coiled-coil domain containing 82 (ENSG00000149231), score: -0.75 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.71 CD82CD82 molecule (ENSG00000085117), score: 0.65 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.83 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: -0.62 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.74 CEPT1choline/ethanolamine phosphotransferase 1 (ENSG00000134255), score: 0.67 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.53 CLTBclathrin, light chain B (ENSG00000175416), score: -0.7 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.79 COX10COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) (ENSG00000006695), score: 0.55 CPSF6cleavage and polyadenylation specific factor 6, 68kDa (ENSG00000111605), score: 0.53 CREB3L3cAMP responsive element binding protein 3-like 3 (ENSG00000060566), score: 0.61 CREG1cellular repressor of E1A-stimulated genes 1 (ENSG00000143162), score: 0.56 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.62 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.93 CTDSPLCTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like (ENSG00000144677), score: 0.58 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.63 CUEDC2CUE domain containing 2 (ENSG00000107874), score: -0.68 CWF19L2CWF19-like 2, cell cycle control (S. pombe) (ENSG00000152404), score: -0.61 CYB5R2cytochrome b5 reductase 2 (ENSG00000166394), score: 0.56 CYLDcylindromatosis (turban tumor syndrome) (ENSG00000083799), score: -0.64 CYP39A1cytochrome P450, family 39, subfamily A, polypeptide 1 (ENSG00000146233), score: 0.62 CYTBcytochrome b (ENSG00000198727), score: -0.92 CYTH1cytohesin 1 (ENSG00000108669), score: -0.8 DCTDdCMP deaminase (ENSG00000129187), score: -0.62 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (ENSG00000109184), score: -0.64 DERL1Der1-like domain family, member 1 (ENSG00000136986), score: 0.67 DERL3Der1-like domain family, member 3 (ENSG00000099958), score: 0.7 DHDPSLdihydrodipicolinate synthase-like, mitochondrial (ENSG00000241935), score: 0.57 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.72 DHX35DEAH (Asp-Glu-Ala-His) box polypeptide 35 (ENSG00000101452), score: -0.65 DLSTdihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) (ENSG00000119689), score: 0.57 EEA1early endosome antigen 1 (ENSG00000102189), score: 0.68 EIF3Deukaryotic translation initiation factor 3, subunit D (ENSG00000100353), score: 0.63 EIF3Meukaryotic translation initiation factor 3, subunit M (ENSG00000149100), score: 0.67 EIF4EBP1eukaryotic translation initiation factor 4E binding protein 1 (ENSG00000187840), score: 0.55 ELMO3engulfment and cell motility 3 (ENSG00000102890), score: 0.52 EME1essential meiotic endonuclease 1 homolog 1 (S. pombe) (ENSG00000154920), score: 0.52 ENTPD6ectonucleoside triphosphate diphosphohydrolase 6 (putative) (ENSG00000197586), score: -0.6 EPHA1EPH receptor A1 (ENSG00000146904), score: 0.58 EPYCepiphycan (ENSG00000083782), score: 0.52 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.82 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.79 ESR1estrogen receptor 1 (ENSG00000091831), score: 0.65 ETFDHelectron-transferring-flavoprotein dehydrogenase (ENSG00000171503), score: 0.55 EXD2exonuclease 3'-5' domain containing 2 (ENSG00000081177), score: -0.77 EXOC3exocyst complex component 3 (ENSG00000180104), score: -0.67 EXOSC9exosome component 9 (ENSG00000123737), score: 0.52 FAM168Bfamily with sequence similarity 168, member B (ENSG00000152102), score: -0.64 FAM172Afamily with sequence similarity 172, member A (ENSG00000113391), score: 0.52 FAM18Afamily with sequence similarity 18, member A (ENSG00000166676), score: -0.68 FAM3Bfamily with sequence similarity 3, member B (ENSG00000183844), score: 0.53 FAM3Dfamily with sequence similarity 3, member D (ENSG00000198643), score: 0.72 FAM82A2family with sequence similarity 82, member A2 (ENSG00000137824), score: 0.79 FAM82Bfamily with sequence similarity 82, member B (ENSG00000176623), score: 0.63 FBXO2F-box protein 2 (ENSG00000116661), score: -0.63 FBXO25F-box protein 25 (ENSG00000147364), score: -0.64 FBXO3F-box protein 3 (ENSG00000110429), score: 0.78 FBXO4F-box protein 4 (ENSG00000151876), score: 0.66 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.55 FETUBfetuin B (ENSG00000090512), score: 0.56 FEZ1fasciculation and elongation protein zeta 1 (zygin I) (ENSG00000149557), score: -0.61 FKBP9FK506 binding protein 9, 63 kDa (ENSG00000122642), score: 0.52 FLVCR1feline leukemia virus subgroup C cellular receptor 1 (ENSG00000162769), score: 0.53 FNDC5fibronectin type III domain containing 5 (ENSG00000160097), score: -0.6 FRA10AC1fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 (ENSG00000148690), score: -0.7 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.62 FZD2frizzled homolog 2 (Drosophila) (ENSG00000180340), score: 0.58 FZD4frizzled homolog 4 (Drosophila) (ENSG00000174804), score: 0.63 FZD5frizzled homolog 5 (Drosophila) (ENSG00000163251), score: 0.52 GALK2galactokinase 2 (ENSG00000156958), score: 0.56 GARSglycyl-tRNA synthetase (ENSG00000106105), score: 0.64 GAS6growth arrest-specific 6 (ENSG00000183087), score: -0.67 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.67 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.76 GGCTgamma-glutamylcyclotransferase (ENSG00000006625), score: 0.6 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: 0.69 GLT8D1glycosyltransferase 8 domain containing 1 (ENSG00000016864), score: -0.6 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: -0.65 GNMTglycine N-methyltransferase (ENSG00000124713), score: 0.53 GOT1glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (ENSG00000120053), score: 0.7 GPBP1L1GC-rich promoter binding protein 1-like 1 (ENSG00000159592), score: 0.53 GPX7glutathione peroxidase 7 (ENSG00000116157), score: 0.56 GRAMD2GRAM domain containing 2 (ENSG00000175318), score: 0.54 GRK6G protein-coupled receptor kinase 6 (ENSG00000198055), score: -0.63 GTPBP8GTP-binding protein 8 (putative) (ENSG00000163607), score: 0.62 HADHAhydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit (ENSG00000084754), score: 0.54 HALhistidine ammonia-lyase (ENSG00000084110), score: 0.53 HM13histocompatibility (minor) 13 (ENSG00000101294), score: 0.62 HMGXB4HMG box domain containing 4 (ENSG00000100281), score: 0.67 IDEinsulin-degrading enzyme (ENSG00000119912), score: 0.55 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.85 IL6STinterleukin 6 signal transducer (gp130, oncostatin M receptor) (ENSG00000134352), score: 0.55 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.73 ITGB1BP1integrin beta 1 binding protein 1 (ENSG00000119185), score: -0.67 IYDiodotyrosine deiodinase (ENSG00000009765), score: 0.58 KBTBD10kelch repeat and BTB (POZ) domain containing 10 (ENSG00000163093), score: -0.6 KBTBD8kelch repeat and BTB (POZ) domain containing 8 (ENSG00000163376), score: 0.53 KCTD6potassium channel tetramerisation domain containing 6 (ENSG00000168301), score: 0.53 KHDRBS3KH domain containing, RNA binding, signal transduction associated 3 (ENSG00000131773), score: -0.66 KIAA0090KIAA0090 (ENSG00000127463), score: 0.7 KIAA0247KIAA0247 (ENSG00000100647), score: 0.53 KIAA0664KIAA0664 (ENSG00000132361), score: 0.67 KIAA1432KIAA1432 (ENSG00000107036), score: 0.8 KIAA1826KIAA1826 (ENSG00000170903), score: -0.63 KIF2Akinesin heavy chain member 2A (ENSG00000068796), score: -0.6 KIFAP3kinesin-associated protein 3 (ENSG00000075945), score: -0.7 KIFC3kinesin family member C3 (ENSG00000140859), score: -0.71 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -1 KSR1kinase suppressor of ras 1 (ENSG00000141068), score: -0.62 LANCL2LanC lantibiotic synthetase component C-like 2 (bacterial) (ENSG00000132434), score: -0.62 LECT2leukocyte cell-derived chemotaxin 2 (ENSG00000145826), score: 0.57 LEPRE1leucine proline-enriched proteoglycan (leprecan) 1 (ENSG00000117385), score: 0.6 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: 0.64 LOC100134387similar to DEAH (Asp-Glu-Ala-His) box polypeptide 40 (ENSG00000108406), score: -0.61 LOC100292021similar to thioredoxin peroxidase (ENSG00000123131), score: 0.53 LRATlecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) (ENSG00000121207), score: 0.58 LY75lymphocyte antigen 75 (ENSG00000054219), score: 0.71 LYSMD3LysM, putative peptidoglycan-binding, domain containing 3 (ENSG00000176018), score: 0.57 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: -0.75 MAGT1magnesium transporter 1 (ENSG00000102158), score: 0.56 MARVELD3MARVEL domain containing 3 (ENSG00000140832), score: 0.53 MCEEmethylmalonyl CoA epimerase (ENSG00000124370), score: 0.54 MCM9minichromosome maintenance complex component 9 (ENSG00000111877), score: -0.63 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (ENSG00000082212), score: -0.74 MFN1mitofusin 1 (ENSG00000171109), score: 0.56 MGAT4Amannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A (ENSG00000071073), score: -0.71 MGST3microsomal glutathione S-transferase 3 (ENSG00000143198), score: 0.57 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.86 MRPS30mitochondrial ribosomal protein S30 (ENSG00000112996), score: 0.68 MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.56 MTERFD1MTERF domain containing 1 (ENSG00000156469), score: 0.64 MTFMTmitochondrial methionyl-tRNA formyltransferase (ENSG00000103707), score: 0.63 MTM1myotubularin 1 (ENSG00000171100), score: 0.52 MTMR14myotubularin related protein 14 (ENSG00000163719), score: 0.76 MTMR2myotubularin related protein 2 (ENSG00000087053), score: -0.68 MTPAPmitochondrial poly(A) polymerase (ENSG00000107951), score: 0.54 MYH10myosin, heavy chain 10, non-muscle (ENSG00000133026), score: -0.79 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.74 NADSYN1NAD synthetase 1 (ENSG00000172890), score: 0.73 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.85 NCSTNnicastrin (ENSG00000162736), score: 0.67 ND2MTND2 (ENSG00000198763), score: -0.9 NDUFA6NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa (ENSG00000184983), score: 0.68 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.71 NEK8NIMA (never in mitosis gene a)- related kinase 8 (ENSG00000160602), score: 0.68 NELFnasal embryonic LHRH factor (ENSG00000165802), score: -0.75 NFIAnuclear factor I/A (ENSG00000162599), score: 0.53 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: 0.54 NIPA1non imprinted in Prader-Willi/Angelman syndrome 1 (ENSG00000170113), score: -0.7 NOD1nucleotide-binding oligomerization domain containing 1 (ENSG00000106100), score: 0.63 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.93 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.53 NRBF2nuclear receptor binding factor 2 (ENSG00000148572), score: 0.77 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.6 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.75 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.63 OSBP2oxysterol binding protein 2 (ENSG00000184792), score: -0.67 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: -0.69 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.59 OTCornithine carbamoyltransferase (ENSG00000036473), score: 0.59 OXSR1oxidative-stress responsive 1 (ENSG00000172939), score: 0.54 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (ENSG00000122884), score: 0.7 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.72 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: -0.81 PACSIN3protein kinase C and casein kinase substrate in neurons 3 (ENSG00000165912), score: 0.55 PAFAH1B2platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) (ENSG00000168092), score: -0.78 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.6 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.65 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (ENSG00000102981), score: -0.68 PARP1poly (ADP-ribose) polymerase 1 (ENSG00000143799), score: -0.62 PCK1phosphoenolpyruvate carboxykinase 1 (soluble) (ENSG00000124253), score: 0.66 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.68 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.52 PDK4pyruvate dehydrogenase kinase, isozyme 4 (ENSG00000004799), score: 0.64 PDP2pyruvate dehyrogenase phosphatase catalytic subunit 2 (ENSG00000172840), score: 0.57 PDZD8PDZ domain containing 8 (ENSG00000165650), score: 0.53 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.64 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.79 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (ENSG00000170525), score: -0.72 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.55 PHF5APHD finger protein 5A (ENSG00000100410), score: 0.59 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.71 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.85 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.73 PODNpodocan (ENSG00000174348), score: 0.71 POLR2Cpolymerase (RNA) II (DNA directed) polypeptide C, 33kDa (ENSG00000102978), score: -0.7 PPEF2protein phosphatase, EF-hand calcium binding domain 2 (ENSG00000156194), score: 0.75 PSEN2presenilin 2 (Alzheimer disease 4) (ENSG00000143801), score: 0.58 PTCH1patched 1 (ENSG00000185920), score: -0.72 PTPRGprotein tyrosine phosphatase, receptor type, G (ENSG00000144724), score: 0.74 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.71 QSER1glutamine and serine rich 1 (ENSG00000060749), score: 0.68 QSOX1quiescin Q6 sulfhydryl oxidase 1 (ENSG00000116260), score: -0.7 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.59 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.67 RASA3RAS p21 protein activator 3 (ENSG00000185989), score: -0.76 RASL11ARAS-like, family 11, member A (ENSG00000122035), score: -0.66 RC3H1ring finger and CCCH-type zinc finger domains 1 (ENSG00000135870), score: 0.57 RCAN1regulator of calcineurin 1 (ENSG00000159200), score: -0.67 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.65 RGS2regulator of G-protein signaling 2, 24kDa (ENSG00000116741), score: -0.75 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.58 RMND1required for meiotic nuclear division 1 homolog (S. cerevisiae) (ENSG00000155906), score: 0.55 RNF103ring finger protein 103 (ENSG00000239305), score: 0.56 RNF144Aring finger protein 144A (ENSG00000151692), score: -0.64 RPS6KA2ribosomal protein S6 kinase, 90kDa, polypeptide 2 (ENSG00000071242), score: -0.6 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.64 RTN4IP1reticulon 4 interacting protein 1 (ENSG00000130347), score: 0.64 SAP30LSAP30-like (ENSG00000164576), score: 0.55 SCAMP2secretory carrier membrane protein 2 (ENSG00000140497), score: 0.52 SCARB1scavenger receptor class B, member 1 (ENSG00000073060), score: 0.58 SDC2syndecan 2 (ENSG00000169439), score: 0.64 SDF2stromal cell-derived factor 2 (ENSG00000132581), score: 0.59 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.64 SERPINA10serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 (ENSG00000140093), score: 0.55 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (ENSG00000135919), score: -0.61 SGPL1sphingosine-1-phosphate lyase 1 (ENSG00000166224), score: 0.61 SGSM2small G protein signaling modulator 2 (ENSG00000141258), score: -0.71 SIN3BSIN3 homolog B, transcription regulator (yeast) (ENSG00000127511), score: -0.68 SLC15A4solute carrier family 15, member 4 (ENSG00000139370), score: 0.74 SLC16A10solute carrier family 16, member 10 (aromatic amino acid transporter) (ENSG00000112394), score: 0.66 SLC16A2solute carrier family 16, member 2 (monocarboxylic acid transporter 8) (ENSG00000147100), score: 0.59 SLC24A3solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 (ENSG00000185052), score: -0.66 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (ENSG00000100372), score: 0.6 SLC25A3solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 (ENSG00000075415), score: 0.59 SLC25A40solute carrier family 25, member 40 (ENSG00000075303), score: 0.77 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.66 SLC38A1solute carrier family 38, member 1 (ENSG00000111371), score: -0.63 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.94 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.74 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.62 SLC7A2solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 (ENSG00000003989), score: 0.62 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (ENSG00000103064), score: -0.64 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (ENSG00000100335), score: 0.61 SNX13sorting nexin 13 (ENSG00000071189), score: 0.71 SNX2sorting nexin 2 (ENSG00000205302), score: 0.63 SNX24sorting nexin 24 (ENSG00000064652), score: 0.73 SPATS2spermatogenesis associated, serine-rich 2 (ENSG00000123352), score: -0.67 SS18synovial sarcoma translocation, chromosome 18 (ENSG00000141380), score: 0.63 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: 0.71 STAB2stabilin 2 (ENSG00000136011), score: 0.69 STAMBPSTAM binding protein (ENSG00000124356), score: 0.56 STK40serine/threonine kinase 40 (ENSG00000196182), score: 0.56 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.79 STX16syntaxin 16 (ENSG00000124222), score: -0.63 SUCLG1succinate-CoA ligase, alpha subunit (ENSG00000163541), score: 0.66 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: 0.61 SYDE2synapse defective 1, Rho GTPase, homolog 2 (C. elegans) (ENSG00000097096), score: 0.55 TALDO1transaldolase 1 (ENSG00000177156), score: 0.52 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (ENSG00000133138), score: 0.65 TCF3transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) (ENSG00000071564), score: 0.76 TCP11L2t-complex 11 (mouse)-like 2 (ENSG00000166046), score: 0.54 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (ENSG00000151923), score: 0.58 TMCO3transmembrane and coiled-coil domains 3 (ENSG00000150403), score: -0.75 TMED5transmembrane emp24 protein transport domain containing 5 (ENSG00000117500), score: 0.58 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.55 TMEM168transmembrane protein 168 (ENSG00000146802), score: 0.54 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.98 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.66 TMEM53transmembrane protein 53 (ENSG00000126106), score: 0.59 TMEM55Atransmembrane protein 55A (ENSG00000155099), score: -0.73 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.86 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.73 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.89 TRAF3TNF receptor-associated factor 3 (ENSG00000131323), score: -0.79 TRAM2translocation associated membrane protein 2 (ENSG00000065308), score: 0.65 TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: 0.55 TRRAPtransformation/transcription domain-associated protein (ENSG00000196367), score: -0.73 TSC22D2TSC22 domain family, member 2 (ENSG00000196428), score: 0.68 TSPAN9tetraspanin 9 (ENSG00000011105), score: 0.53 TTC36tetratricopeptide repeat domain 36 (ENSG00000172425), score: 0.55 TTF2transcription termination factor, RNA polymerase II (ENSG00000116830), score: 0.76 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.77 TWISTNBTWIST neighbor (ENSG00000105849), score: 0.69 TWSG1twisted gastrulation homolog 1 (Drosophila) (ENSG00000128791), score: -0.68 TXNDC12thioredoxin domain containing 12 (endoplasmic reticulum) (ENSG00000117862), score: 0.71 TXNRD1thioredoxin reductase 1 (ENSG00000198431), score: 0.52 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.74 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.6 UCK1uridine-cytidine kinase 1 (ENSG00000130717), score: 0.57 UNC119unc-119 homolog (C. elegans) (ENSG00000109103), score: -0.61 UPB1ureidopropionase, beta (ENSG00000100024), score: 0.53 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: 0.62 UTP11LUTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) (ENSG00000183520), score: 0.58 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.58 VAC14Vac14 homolog (S. cerevisiae) (ENSG00000103043), score: 0.54 VPS39vacuolar protein sorting 39 homolog (S. cerevisiae) (ENSG00000166887), score: -0.77 WDR37WD repeat domain 37 (ENSG00000047056), score: -0.65 WDR43WD repeat domain 43 (ENSG00000163811), score: 0.69 WDR6WD repeat domain 6 (ENSG00000178252), score: -0.64 XKR9XK, Kell blood group complex subunit-related family, member 9 (ENSG00000221947), score: 0.66 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: 0.53 YIPF1Yip1 domain family, member 1 (ENSG00000058799), score: 0.52 ZBED4zinc finger, BED-type containing 4 (ENSG00000100426), score: 0.53 ZBTB11zinc finger and BTB domain containing 11 (ENSG00000066422), score: -0.68 ZBTB2zinc finger and BTB domain containing 2 (ENSG00000181472), score: 0.62 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.83 ZDHHC13zinc finger, DHHC-type containing 13 (ENSG00000177054), score: 0.59 ZDHHC21zinc finger, DHHC-type containing 21 (ENSG00000175893), score: -0.69 ZNF276zinc finger protein 276 (ENSG00000158805), score: 0.62 ZNF346zinc finger protein 346 (ENSG00000113761), score: -0.63 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.53 ZRANB3zinc finger, RAN-binding domain containing 3 (ENSG00000121988), score: -0.62 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.64
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |