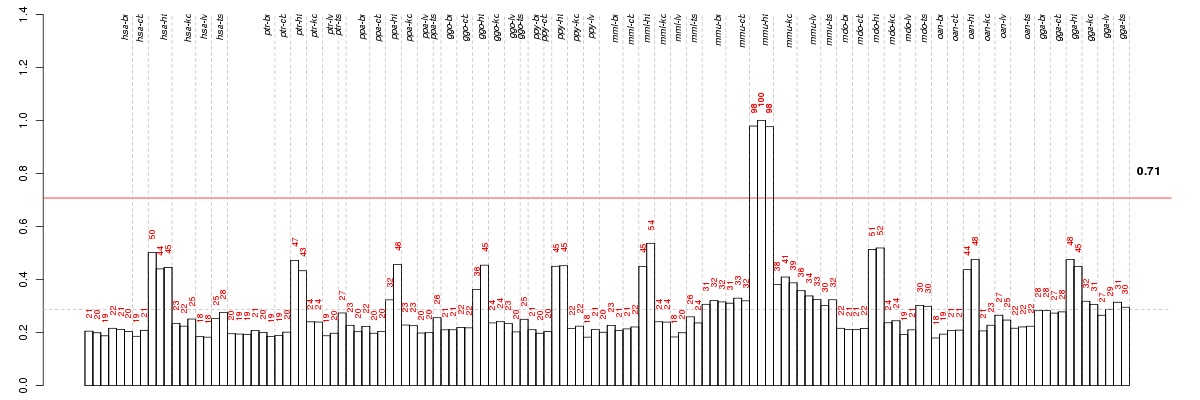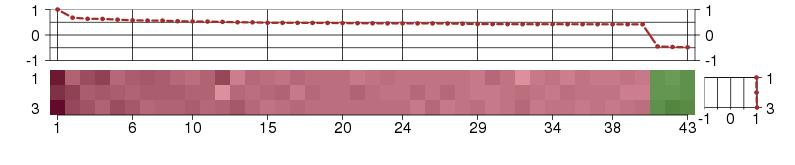

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
sarcolemma
The outer membrane of a muscle cell, consisting of the plasma membrane, a covering basement membrane (about 100 nm thick and sometimes common to more than one fiber), and the associated loose network of collagen fibers.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

ANGPT1angiopoietin 1 (ENSG00000154188), score: 0.43 ASB11ankyrin repeat and SOCS box-containing 11 (ENSG00000165192), score: 0.53 ASB14ankyrin repeat and SOCS box-containing 14 (ENSG00000239388), score: 0.43 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (ENSG00000174437), score: 0.5 CACNA1Scalcium channel, voltage-dependent, L type, alpha 1S subunit (ENSG00000081248), score: 0.6 CADM1cell adhesion molecule 1 (ENSG00000182985), score: -0.47 CD28CD28 molecule (ENSG00000178562), score: 0.42 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.46 CHRNA10cholinergic receptor, nicotinic, alpha 10 (ENSG00000129749), score: 0.45 CYTL1cytokine-like 1 (ENSG00000170891), score: 0.63 EGLN1egl nine homolog 1 (C. elegans) (ENSG00000135766), score: 0.46 EHD4EH-domain containing 4 (ENSG00000103966), score: 0.45 FAM160A1family with sequence similarity 160, member A1 (ENSG00000164142), score: 0.45 FAM174Bfamily with sequence similarity 174, member B (ENSG00000185442), score: 0.53 H1FXH1 histone family, member X (ENSG00000184897), score: -0.48 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.42 IVNS1ABPinfluenza virus NS1A binding protein (ENSG00000116679), score: 0.48 KCNJ5potassium inwardly-rectifying channel, subfamily J, member 5 (ENSG00000120457), score: 0.48 KCNV2potassium channel, subfamily V, member 2 (ENSG00000168263), score: 0.68 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.42 LMOD3leiomodin 3 (fetal) (ENSG00000163380), score: 0.56 LRRC10leucine rich repeat containing 10 (ENSG00000198812), score: 0.45 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.48 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 1 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (ENSG00000160285), score: -0.45 MFN1mitofusin 1 (ENSG00000171109), score: 0.46 MYL1myosin, light chain 1, alkali; skeletal, fast (ENSG00000168530), score: 0.57 MYLK3myosin light chain kinase 3 (ENSG00000140795), score: 0.44 MYLK4myosin light chain kinase family, member 4 (ENSG00000145949), score: 0.64 MYOCDmyocardin (ENSG00000141052), score: 0.45 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.42 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (ENSG00000105953), score: 0.47 PHTF2putative homeodomain transcription factor 2 (ENSG00000006576), score: 0.58 PPP1R3Aprotein phosphatase 1, regulatory (inhibitor) subunit 3A (ENSG00000154415), score: 0.42 RAPSNreceptor-associated protein of the synapse (ENSG00000165917), score: 0.54 RNF139ring finger protein 139 (ENSG00000170881), score: 0.42 SGCGsarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) (ENSG00000102683), score: 0.43 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.42 TEAD1TEA domain family member 1 (SV40 transcriptional enhancer factor) (ENSG00000187079), score: 0.42 TXLNBtaxilin beta (ENSG00000164440), score: 0.47 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.42 UCP3uncoupling protein 3 (mitochondrial, proton carrier) (ENSG00000175564), score: 0.51 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.5
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |