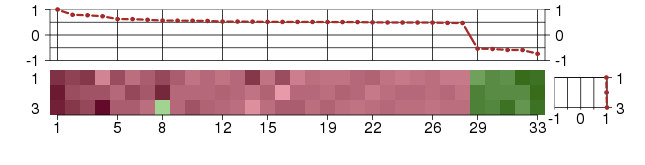

Under-expression is coded with green,
over-expression with red color.



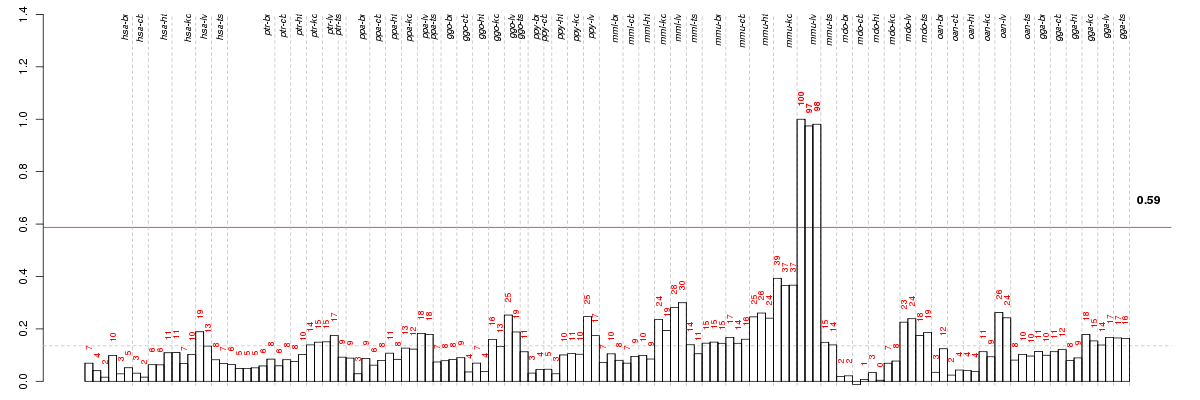
ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.51 AQP8aquaporin 8 (ENSG00000103375), score: 0.56 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.55 BBS2Bardet-Biedl syndrome 2 (ENSG00000125124), score: -0.59 C12orf72chromosome 12 open reading frame 72 (ENSG00000139160), score: 0.51 C2CD2C2 calcium-dependent domain containing 2 (ENSG00000157617), score: 0.49 C7orf23chromosome 7 open reading frame 23 (ENSG00000135185), score: 0.55 CCBL2cysteine conjugate-beta lyase 2 (ENSG00000137944), score: 0.52 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.51 CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.52 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.79 DNTTdeoxynucleotidyltransferase, terminal (ENSG00000107447), score: 0.62 FABP2fatty acid binding protein 2, intestinal (ENSG00000145384), score: 1 GJB2gap junction protein, beta 2, 26kDa (ENSG00000165474), score: 0.49 GNEglucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase (ENSG00000159921), score: 0.51 GPLD1glycosylphosphatidylinositol specific phospholipase D1 (ENSG00000112293), score: 0.47 H1FXH1 histone family, member X (ENSG00000184897), score: -0.53 LIFRleukemia inhibitory factor receptor alpha (ENSG00000113594), score: 0.6 LRIT2leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 (ENSG00000204033), score: 0.57 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.62 MIA3melanoma inhibitory activity family, member 3 (ENSG00000154305), score: 0.51 PLK3polo-like kinase 3 (ENSG00000173846), score: 0.56 PRKD3protein kinase D3 (ENSG00000115825), score: 0.53 PRLRprolactin receptor (ENSG00000113494), score: 0.49 SCCPDHsaccharopine dehydrogenase (putative) (ENSG00000143653), score: -0.59 SDR42E1short chain dehydrogenase/reductase family 42E, member 1 (ENSG00000184860), score: 0.53 SLC17A8solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 (ENSG00000179520), score: 0.74 SNTG2syntrophin, gamma 2 (ENSG00000172554), score: 0.47 TSPAN3tetraspanin 3 (ENSG00000140391), score: -0.74 TTC39Ctetratricopeptide repeat domain 39C (ENSG00000168234), score: 0.5 XKR9XK, Kell blood group complex subunit-related family, member 9 (ENSG00000221947), score: 0.49 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.51 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.77
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |