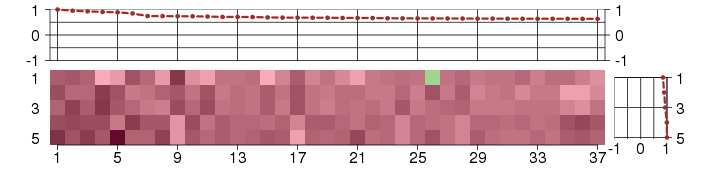

Under-expression is coded with green,
over-expression with red color.



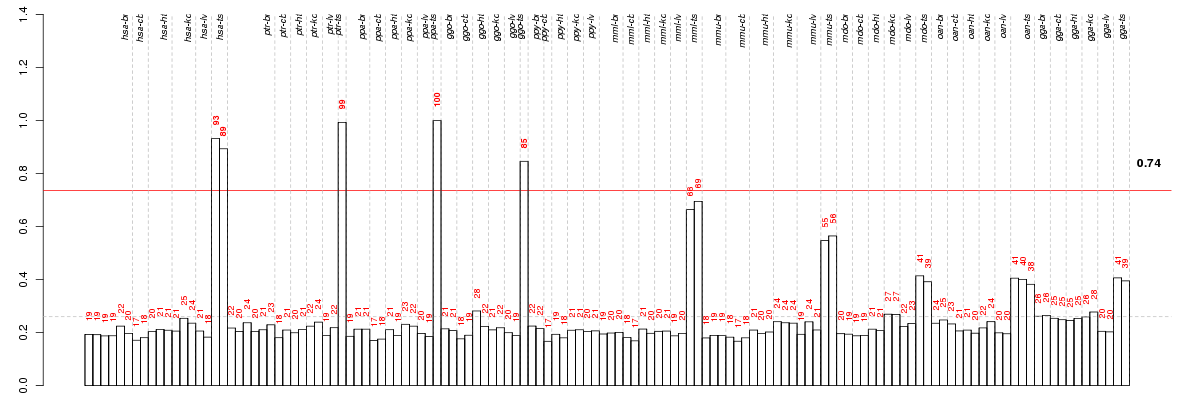
ARHGAP28Rho GTPase activating protein 28 (ENSG00000088756), score: 0.67 BARX1BARX homeobox 1 (ENSG00000131668), score: 0.74 BOLLbol, boule-like (Drosophila) (ENSG00000152430), score: 0.64 BRIP1BRCA1 interacting protein C-terminal helicase 1 (ENSG00000136492), score: 0.64 BTG4B-cell translocation gene 4 (ENSG00000137707), score: 0.71 C20orf151chromosome 20 open reading frame 151 (ENSG00000130701), score: 0.67 CLSPNclaspin (ENSG00000092853), score: 0.64 DEF6differentially expressed in FDCP 6 homolog (mouse) (ENSG00000023892), score: 0.64 DMBX1diencephalon/mesencephalon homeobox 1 (ENSG00000197587), score: 0.72 DNAJC5BDnaJ (Hsp40) homolog, subfamily C, member 5 beta (ENSG00000147570), score: 0.64 DRG1developmentally regulated GTP binding protein 1 (ENSG00000185721), score: 0.73 DTLdenticleless homolog (Drosophila) (ENSG00000143476), score: 0.65 EVX1even-skipped homeobox 1 (ENSG00000106038), score: 0.89 GJA8gap junction protein, alpha 8, 50kDa (ENSG00000121634), score: 0.93 GLRA4glycine receptor, alpha 4 (ENSG00000188828), score: 0.65 GRHL3grainyhead-like 3 (Drosophila) (ENSG00000158055), score: 0.65 GRXCR2glutaredoxin, cysteine rich 2 (ENSG00000204928), score: 0.63 GSG1germ cell associated 1 (ENSG00000111305), score: 0.71 HSF2BPheat shock transcription factor 2 binding protein (ENSG00000160207), score: 0.66 IGF2BP1insulin-like growth factor 2 mRNA binding protein 1 (ENSG00000159217), score: 0.65 KCNV2potassium channel, subfamily V, member 2 (ENSG00000168263), score: 0.65 KRT23keratin 23 (histone deacetylase inducible) (ENSG00000108244), score: 0.68 LAMP3lysosomal-associated membrane protein 3 (ENSG00000078081), score: 0.69 LASS3LAG1 homolog, ceramide synthase 3 (ENSG00000154227), score: 0.68 LOC81691exonuclease NEF-sp (ENSG00000005189), score: 0.7 LOXHD1lipoxygenase homology domains 1 (ENSG00000167210), score: 0.95 LRIT1leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 (ENSG00000148602), score: 1 OPN5opsin 5 (ENSG00000124818), score: 0.91 PPM1Gprotein phosphatase, Mg2+/Mn2+ dependent, 1G (ENSG00000115241), score: 0.68 PROK1prokineticin 1 (ENSG00000143125), score: 0.74 PRTGprotogenin (ENSG00000166450), score: 0.85 RAE1RAE1 RNA export 1 homolog (S. pombe) (ENSG00000101146), score: 0.74 SMURF1SMAD specific E3 ubiquitin protein ligase 1 (ENSG00000198742), score: 0.64 SPSB4splA/ryanodine receptor domain and SOCS box containing 4 (ENSG00000175093), score: 0.68 TTC29tetratricopeptide repeat domain 29 (ENSG00000137473), score: 0.64 VPRBPVpr (HIV-1) binding protein (ENSG00000145041), score: 0.63 VRK3vaccinia related kinase 3 (ENSG00000105053), score: 0.67
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_ts_m_ca1 | ggo | ts | m | _ |
| hsa_ts_m1_ca1 | hsa | ts | m | 1 |
| hsa_ts_m2_ca1 | hsa | ts | m | 2 |
| ptr_ts_m_ca1 | ptr | ts | m | _ |
| ppa_ts_m_ca1 | ppa | ts | m | _ |