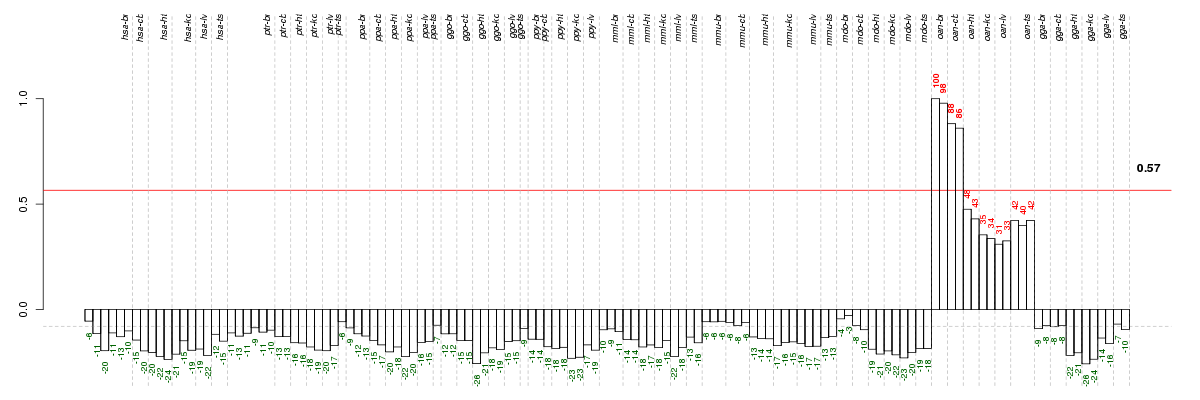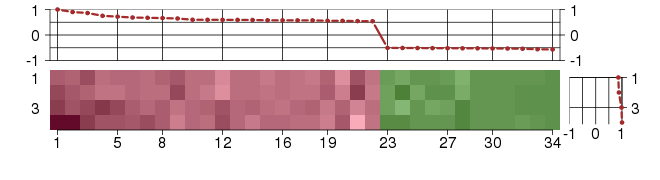

Under-expression is coded with green,
over-expression with red color.



AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.67 AVPR1Barginine vasopressin receptor 1B (ENSG00000198049), score: 1 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.51 C1orf25chromosome 1 open reading frame 25 (ENSG00000121486), score: 0.59 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: 0.58 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.51 CHRNB4cholinergic receptor, nicotinic, beta 4 (ENSG00000117971), score: 0.55 CNTN5contactin 5 (ENSG00000149972), score: 0.72 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -0.57 CPLX4complexin 4 (ENSG00000166569), score: 0.75 DCTN4dynactin 4 (p62) (ENSG00000132912), score: 0.6 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.57 EXTL2exostoses (multiple)-like 2 (ENSG00000162694), score: 0.57 FLNBfilamin B, beta (ENSG00000136068), score: -0.52 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.52 GUCY2Cguanylate cyclase 2C (heat stable enterotoxin receptor) (ENSG00000070019), score: 0.59 HHLA1HERV-H LTR-associating 1 (ENSG00000132297), score: 0.86 IL12RB2interleukin 12 receptor, beta 2 (ENSG00000081985), score: 0.56 LOC100293905similar to gastrin-releasing peptide receptor (ENSG00000126010), score: 0.65 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.69 MLXIPMLX interacting protein (ENSG00000175727), score: -0.52 PRPH2peripherin 2 (retinal degeneration, slow) (ENSG00000112619), score: 0.58 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: 0.54 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: -0.56 RPS6KC1ribosomal protein S6 kinase, 52kDa, polypeptide 1 (ENSG00000136643), score: 0.6 SLC35A5solute carrier family 35, member A5 (ENSG00000138459), score: -0.54 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.53 THOC7THO complex 7 homolog (Drosophila) (ENSG00000163634), score: -0.51 TMEM117transmembrane protein 117 (ENSG00000139173), score: 0.59 TRPA1transient receptor potential cation channel, subfamily A, member 1 (ENSG00000104321), score: 0.56 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.52 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.66 WNT3Awingless-type MMTV integration site family, member 3A (ENSG00000154342), score: 0.9 YTHDC2YTH domain containing 2 (ENSG00000047188), score: -0.53
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| oan_br_m_ca1 | oan | br | m | _ |