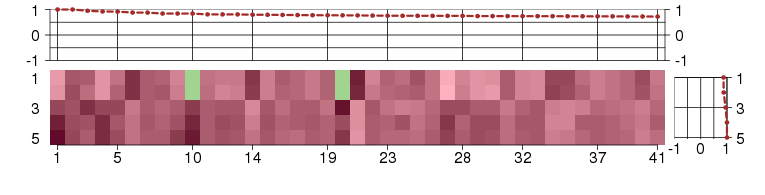

Under-expression is coded with green,
over-expression with red color.



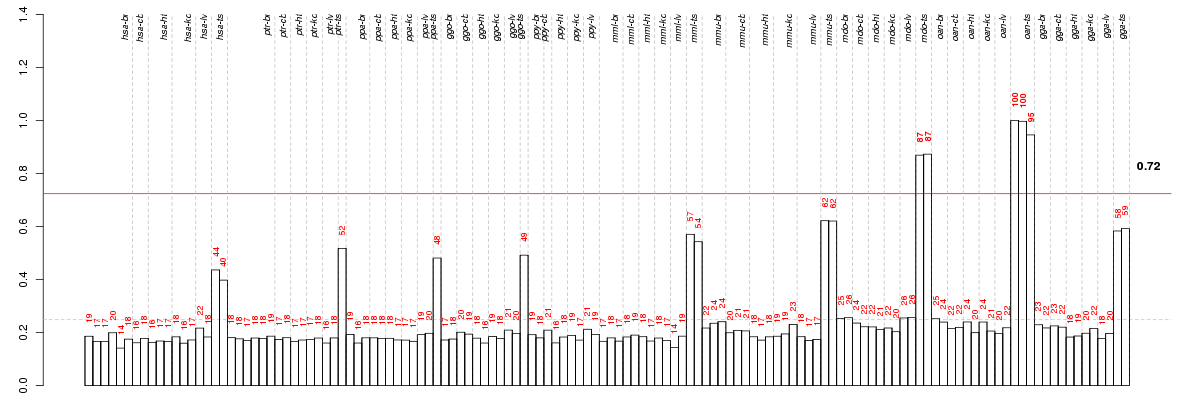
APTXaprataxin (ENSG00000137074), score: 0.8 ATAD2ATPase family, AAA domain containing 2 (ENSG00000156802), score: 0.75 C12orf48chromosome 12 open reading frame 48 (ENSG00000185480), score: 0.81 C14orf39chromosome 14 open reading frame 39 (ENSG00000179008), score: 0.74 C17orf71chromosome 17 open reading frame 71 (ENSG00000167447), score: 0.76 C1orf174chromosome 1 open reading frame 174 (ENSG00000198912), score: 0.84 C1orf9chromosome 1 open reading frame 9 (ENSG00000094975), score: 0.84 CENPIcentromere protein I (ENSG00000102384), score: 0.76 CNGA2cyclic nucleotide gated channel alpha 2 (ENSG00000183862), score: 0.92 CYP11A1cytochrome P450, family 11, subfamily A, polypeptide 1 (ENSG00000140459), score: 0.75 ECDecdysoneless homolog (Drosophila) (ENSG00000122882), score: 0.77 EFCAB1EF-hand calcium binding domain 1 (ENSG00000034239), score: 0.73 FAM54Afamily with sequence similarity 54, member A (ENSG00000146410), score: 0.73 FANCBFanconi anemia, complementation group B (ENSG00000181544), score: 1 GDPD4glycerophosphodiester phosphodiesterase domain containing 4 (ENSG00000178795), score: 0.88 GEMC1geminin coiled-coil domain-containing protein 1 (ENSG00000205835), score: 0.73 GRXCR1glutaredoxin, cysteine rich 1 (ENSG00000215203), score: 1 HORMAD2HORMA domain containing 2 (ENSG00000176635), score: 0.79 KIF14kinesin family member 14 (ENSG00000118193), score: 0.75 LCORLligand dependent nuclear receptor corepressor-like (ENSG00000178177), score: 0.81 LCTlactase (ENSG00000115850), score: 0.95 LHCGRluteinizing hormone/choriogonadotropin receptor (ENSG00000138039), score: 0.92 LRRC18leucine rich repeat containing 18 (ENSG00000165383), score: 0.73 LRRC52leucine rich repeat containing 52 (ENSG00000162763), score: 0.74 LRRIQ4leucine-rich repeats and IQ motif containing 4 (ENSG00000188306), score: 0.72 MTF2metal response element binding transcription factor 2 (ENSG00000143033), score: 0.81 NR5A1nuclear receptor subfamily 5, group A, member 1 (ENSG00000136931), score: 0.78 ORC3Lorigin recognition complex, subunit 3-like (yeast) (ENSG00000135336), score: 0.74 PHTF1putative homeodomain transcription factor 1 (ENSG00000116793), score: 0.73 RAD54BRAD54 homolog B (S. cerevisiae) (ENSG00000197275), score: 0.79 RBP2retinol binding protein 2, cellular (ENSG00000114113), score: 0.77 SGOL1shugoshin-like 1 (S. pombe) (ENSG00000129810), score: 0.88 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (ENSG00000087916), score: 0.84 SMC1Bstructural maintenance of chromosomes 1B (ENSG00000077935), score: 0.75 STILSCL/TAL1 interrupting locus (ENSG00000123473), score: 0.74 STRA8stimulated by retinoic acid gene 8 homolog (mouse) (ENSG00000146857), score: 0.78 TBX4T-box 4 (ENSG00000121075), score: 0.75 TEX9testis expressed 9 (ENSG00000151575), score: 0.73 USP49ubiquitin specific peptidase 49 (ENSG00000164663), score: 0.77 WDR64WD repeat domain 64 (ENSG00000162843), score: 0.74 ZNF438zinc finger protein 438 (ENSG00000183621), score: 0.74
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| oan_ts_m2_ca1 | oan | ts | m | 2 |
| oan_ts_m1_ca1 | oan | ts | m | 1 |
| oan_ts_m3_ca1 | oan | ts | m | 3 |