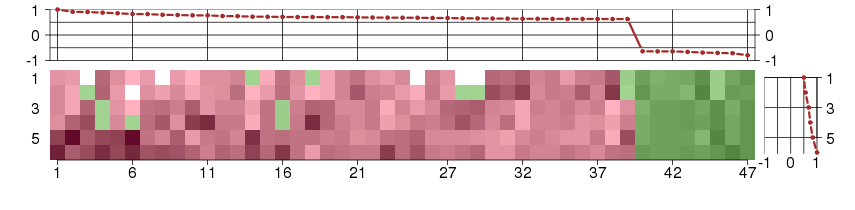

Under-expression is coded with green,
over-expression with red color.

response to tumor cell
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a tumor cell.
regulation of response to biotic stimulus
Any process that modulates the frequency, rate, or extent of a response to biotic stimulus.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to biotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a biotic stimulus, a stimulus caused or produced by a living organism.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
NA
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of response to biotic stimulus
Any process that modulates the frequency, rate, or extent of a response to biotic stimulus.


| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05330 | 1.614e-03 | 0.06333 | 3 | 5 | Allograft rejection |
| 04672 | 2.563e-02 | 0.1773 | 3 | 14 | Intestinal immune network for IgA production |
ABCA12ATP-binding cassette, sub-family A (ABC1), member 12 (ENSG00000144452), score: 0.88 AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.68 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (ENSG00000113108), score: -0.8 C13orf39chromosome 13 open reading frame 39 (ENSG00000139780), score: 0.7 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.72 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.82 CD28CD28 molecule (ENSG00000178562), score: 0.68 CD40LGCD40 ligand (ENSG00000102245), score: 0.66 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 0.79 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.64 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: -0.64 EREGepiregulin (ENSG00000124882), score: 0.77 ESR2estrogen receptor 2 (ER beta) (ENSG00000140009), score: 0.7 FAM109Bfamily with sequence similarity 109, member B (ENSG00000177096), score: 0.63 FBXL18F-box and leucine-rich repeat protein 18 (ENSG00000155034), score: 0.7 FNDC7fibronectin type III domain containing 7 (ENSG00000143107), score: 0.64 HPGDShematopoietic prostaglandin D synthase (ENSG00000163106), score: 0.9 ICOSinducible T-cell co-stimulator (ENSG00000163600), score: 0.77 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.91 IL22RA2interleukin 22 receptor, alpha 2 (ENSG00000164485), score: 0.82 IMPG2interphotoreceptor matrix proteoglycan 2 (ENSG00000081148), score: 0.64 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.66 LCMT1leucine carboxyl methyltransferase 1 (ENSG00000205629), score: -0.71 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.72 PANX3pannexin 3 (ENSG00000154143), score: 0.74 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.64 PDX1pancreatic and duodenal homeobox 1 (ENSG00000139515), score: 0.7 PLRG1pleiotropic regulator 1 (PRL1 homolog, Arabidopsis) (ENSG00000171566), score: -0.69 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (ENSG00000100479), score: 0.67 PRKAG3protein kinase, AMP-activated, gamma 3 non-catalytic subunit (ENSG00000115592), score: 0.65 RPIAribose 5-phosphate isomerase A (ENSG00000153574), score: 0.78 SEPT2septin 2 (ENSG00000168385), score: -0.64 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.85 SPINK4serine peptidase inhibitor, Kazal type 4 (ENSG00000122711), score: 1 TAAR1trace amine associated receptor 1 (ENSG00000146399), score: 0.68 TAF3TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa (ENSG00000165632), score: 0.63 TM7SF4transmembrane 7 superfamily member 4 (ENSG00000164935), score: 0.63 TMEM199transmembrane protein 199 (ENSG00000244045), score: 0.69 TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.65 TNFRSF11Atumor necrosis factor receptor superfamily, member 11a, NFKB activator (ENSG00000141655), score: 0.74 TP63tumor protein p63 (ENSG00000073282), score: 0.63 TRPA1transient receptor potential cation channel, subfamily A, member 1 (ENSG00000104321), score: 0.71 WDR41WD repeat domain 41 (ENSG00000164253), score: -0.7 XPR1xenotropic and polytropic retrovirus receptor 1 (ENSG00000143324), score: 0.63 ZBTB8Bzinc finger and BTB domain containing 8B (ENSG00000215897), score: 0.69 ZNF704zinc finger protein 704 (ENSG00000164684), score: 0.67
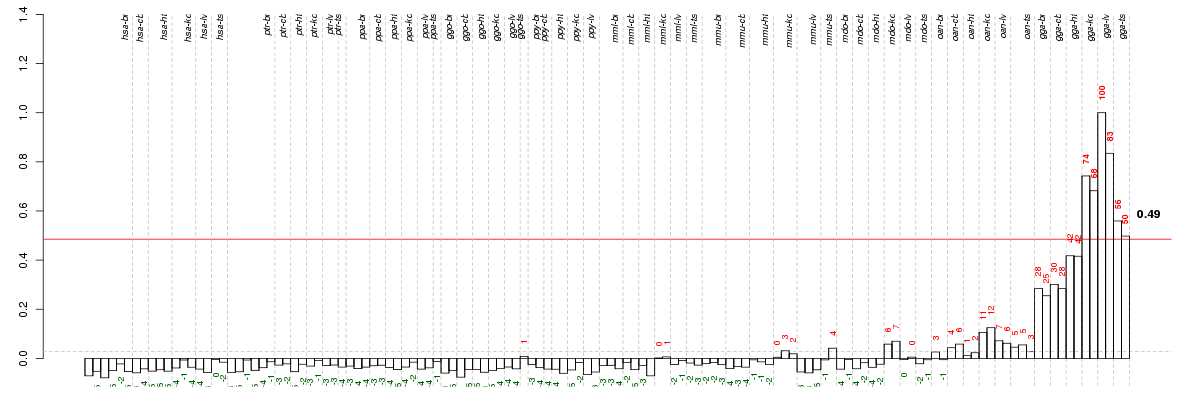
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_ts_m2_ca1 | gga | ts | m | 2 |
| gga_ts_m1_ca1 | gga | ts | m | 1 |
| gga_kd_f_ca1 | gga | kd | f | _ |
| gga_kd_m_ca1 | gga | kd | m | _ |
| gga_lv_f_ca1 | gga | lv | f | _ |
| gga_lv_m_ca1 | gga | lv | m | _ |