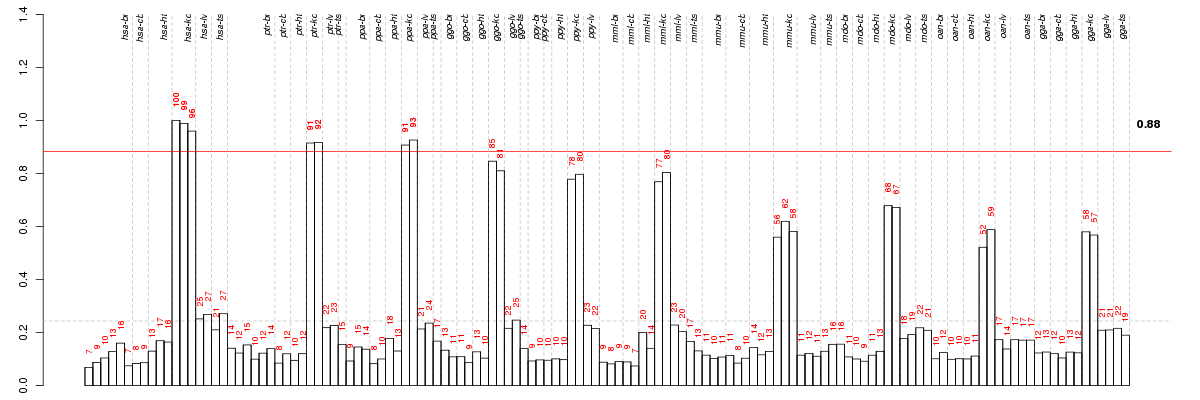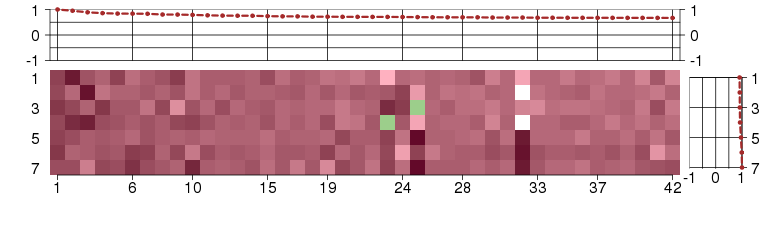

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
secretion
The controlled release of a substance by a cell, a group of cells, or a tissue.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
NA
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
sodium channel complex
An ion channel complex through which sodium ions pass.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
apical part of cell
The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
metallopeptidase activity
Catalysis of the hydrolysis of peptide bonds by a mechanism in which water acts as a nucleophile, one or two metal ions hold the water molecule in place, and charged amino acid side chains are ligands for the metal ions.
peptidase activity
Catalysis of the hydrolysis of a peptide bond. A peptide bond is a covalent bond formed when the carbon atom from the carboxyl group of one amino acid shares electrons with the nitrogen atom from the amino group of a second amino acid.
peptidase activity, acting on L-amino acid peptides
Catalysis of the hydrolysis of peptide bonds formed between L-amino acids.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04960 | 6.242e-04 | 0.1351 | 4 | 16 | Aldosterone-regulated sodium reabsorption |
ACPPacid phosphatase, prostate (ENSG00000014257), score: 0.84 ATP6V0A4ATPase, H+ transporting, lysosomal V0 subunit a4 (ENSG00000105929), score: 0.67 BARX2BARX homeobox 2 (ENSG00000043039), score: 0.8 CA12carbonic anhydrase XII (ENSG00000074410), score: 0.73 CRYAAcrystallin, alpha A (ENSG00000160202), score: 1 DPEP1dipeptidase 1 (renal) (ENSG00000015413), score: 0.67 ELF5E74-like factor 5 (ets domain transcription factor) (ENSG00000135374), score: 0.8 F2RL1coagulation factor II (thrombin) receptor-like 1 (ENSG00000164251), score: 0.69 FOXI1forkhead box I1 (ENSG00000168269), score: 0.76 GATA3GATA binding protein 3 (ENSG00000107485), score: 0.69 GPR114G protein-coupled receptor 114 (ENSG00000159618), score: 0.68 GRHL2grainyhead-like 2 (Drosophila) (ENSG00000083307), score: 0.67 HOXA10homeobox A10 (ENSG00000153807), score: 0.74 HSD11B2hydroxysteroid (11-beta) dehydrogenase 2 (ENSG00000176387), score: 0.68 IL1RL1interleukin 1 receptor-like 1 (ENSG00000115602), score: 0.79 ILDR1immunoglobulin-like domain containing receptor 1 (ENSG00000145103), score: 0.68 IMPA2inositol(myo)-1(or 4)-monophosphatase 2 (ENSG00000141401), score: 0.69 KCNJ1potassium inwardly-rectifying channel, subfamily J, member 1 (ENSG00000151704), score: 0.76 LGALS2lectin, galactoside-binding, soluble, 2 (ENSG00000100079), score: 0.67 LMX1BLIM homeobox transcription factor 1, beta (ENSG00000136944), score: 0.86 MMP7matrix metallopeptidase 7 (matrilysin, uterine) (ENSG00000137673), score: 0.95 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.71 NPHS2nephrosis 2, idiopathic, steroid-resistant (podocin) (ENSG00000116218), score: 0.84 OVCH2ovochymase 2 (gene/pseudogene) (ENSG00000183378), score: 0.89 PAPPA2pappalysin 2 (ENSG00000116183), score: 0.7 PEPDpeptidase D (ENSG00000124299), score: 0.68 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (ENSG00000197943), score: 0.71 PROM2prominin 2 (ENSG00000155066), score: 0.77 RHCGRh family, C glycoprotein (ENSG00000140519), score: 0.72 SCNN1Asodium channel, nonvoltage-gated 1 alpha (ENSG00000111319), score: 0.69 SCNN1Bsodium channel, nonvoltage-gated 1, beta (ENSG00000168447), score: 0.68 SIM2single-minded homolog 2 (Drosophila) (ENSG00000159263), score: 0.72 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (ENSG00000088386), score: 0.71 SLC26A7solute carrier family 26, member 7 (ENSG00000147606), score: 0.71 SLC5A12solute carrier family 5 (sodium/glucose cotransporter), member 12 (ENSG00000148942), score: 0.67 STK32Bserine/threonine kinase 32B (ENSG00000152953), score: 0.73 TFCP2L1transcription factor CP2-like 1 (ENSG00000115112), score: 0.76 TMPRSS4transmembrane protease, serine 4 (ENSG00000137648), score: 0.83 TRPV4transient receptor potential cation channel, subfamily V, member 4 (ENSG00000111199), score: 0.68 VCAM1vascular cell adhesion molecule 1 (ENSG00000162692), score: 0.67 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.71 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.67
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppa_kd_m_ca1 | ppa | kd | m | _ |
| ptr_kd_m_ca1 | ptr | kd | m | _ |
| ptr_kd_f_ca1 | ptr | kd | f | _ |
| ppa_kd_f_ca1 | ppa | kd | f | _ |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |