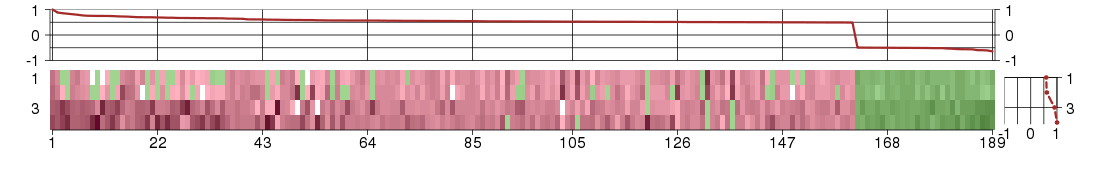

Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
aggressive behavior
A behavioral interaction between organisms in which one organism has the intention of inflicting physical damage on another individual.
maternal aggressive behavior
Aggressive behavior of a female to protect her offspring from a threat.
behavior
The specific actions or reactions of an organism in response to external or internal stimuli. Patterned activity of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions.
mating behavior
The behavioral interactions between organisms for the purpose of mating, or sexual reproduction resulting in the formation of zygotes.
mating
The pairwise union of individuals for the purpose of sexual reproduction, ultimately resulting in the formation of zygotes.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
reproductive behavior
The specific actions or reactions of an organism that are associated with reproduction.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
reproductive behavior in a multicellular organism
The specific actions or reactions of an organism that are associated with reproduction in a multicellular organism.
regulation of female receptivity
Any process that modulates the frequency, rate or extent of the willingness or readiness of a female to receive male advances.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
multi-organism process
Any process by which an organism has an effect on another organism of the same or different species.
behavioral interaction between organisms
Any process by which an organism has a behavioral effect on another organism of the same or different species.
female mating behavior
The specific actions or reactions of a female organism that are associated with reproduction.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
all
NA
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
mating
The pairwise union of individuals for the purpose of sexual reproduction, ultimately resulting in the formation of zygotes.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
mating behavior
The behavioral interactions between organisms for the purpose of mating, or sexual reproduction resulting in the formation of zygotes.
reproductive behavior in a multicellular organism
The specific actions or reactions of an organism that are associated with reproduction in a multicellular organism.
reproductive behavior
The specific actions or reactions of an organism that are associated with reproduction.
behavioral interaction between organisms
Any process by which an organism has a behavioral effect on another organism of the same or different species.
mating behavior
The behavioral interactions between organisms for the purpose of mating, or sexual reproduction resulting in the formation of zygotes.
female mating behavior
The specific actions or reactions of a female organism that are associated with reproduction.
regulation of female receptivity
Any process that modulates the frequency, rate or extent of the willingness or readiness of a female to receive male advances.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
photoreceptor outer segment
The outer segment of a vertebrate photoreceptor that contains discs of photoreceptive membranes.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cilium
A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
nonmotile primary cilium
An immotile primary cilium that may be missing the central pair of microtubules, or the central pair of microtubules and outer dynein arms. Some primary cilia also have altered arrangements of outer microtubules (fewer than nine and/or not always present as doublets). Nonmotile primary cilia typically function as sensory organelles that concentrate and organize sensory signaling molecules.
photoreceptor outer segment membrane
The membrane surrounding the outer segment of a vertebrate photoreceptor.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
photoreceptor outer segment membrane
The membrane surrounding the outer segment of a vertebrate photoreceptor.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
photoreceptor outer segment membrane
The membrane surrounding the outer segment of a vertebrate photoreceptor.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
N-acetylgalactosamine-4-sulfatase activity
Catalysis of the hydrolysis of the 4-sulfate groups of the N-acetyl-D-galactosamine 4-sulfate units of chondroitin sulfate and dermatan sulfate.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
sulfuric ester hydrolase activity
Catalysis of the reaction: RSO-R' + H2O = RSOOH + R'H. This reaction is the hydrolysis of any sulfuric ester bond, any ester formed from sulfuric acid, O=SO(OH)2.
vitamin transporter activity
Enables the directed movement of vitamins into, out of, within or between cells. A vitamin is one of a number of unrelated organic substances that occur in many foods in small amounts and that are necessary in trace amounts for the normal metabolic functioning of the body.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on ester bonds
Catalysis of the hydrolysis of any ester bond.
all
NA
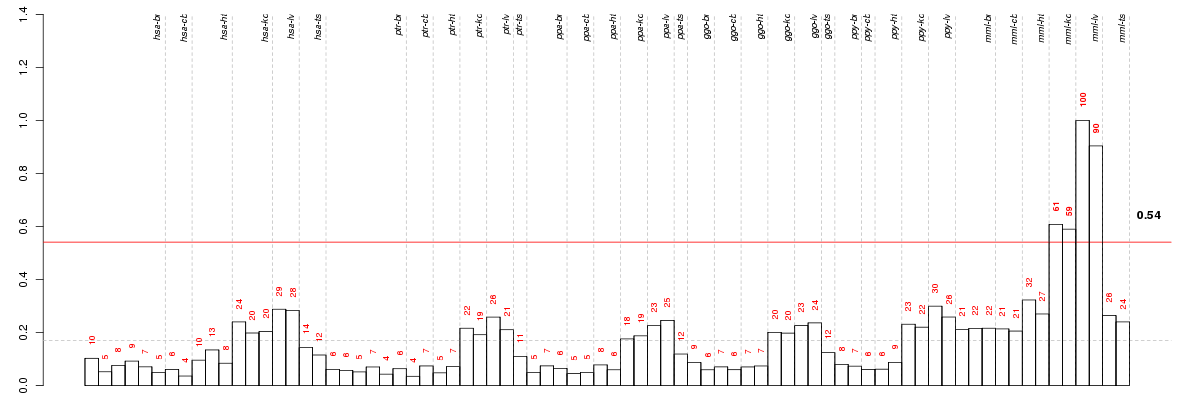
A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.66 ABCA4ATP-binding cassette, sub-family A (ABC1), member 4 (ENSG00000198691), score: 0.68 ABP1amiloride binding protein 1 (amine oxidase (copper-containing)) (ENSG00000002726), score: 0.56 ACCS1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) (ENSG00000110455), score: 0.49 ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.54 ACOX1acyl-CoA oxidase 1, palmitoyl (ENSG00000161533), score: 0.53 ADAM12ADAM metallopeptidase domain 12 (ENSG00000148848), score: 0.88 AFPalpha-fetoprotein (ENSG00000081051), score: 0.81 AGPHD1aminoglycoside phosphotransferase domain containing 1 (ENSG00000188266), score: 0.51 AGR3anterior gradient homolog 3 (Xenopus laevis) (ENSG00000173467), score: 0.52 AIM1Labsent in melanoma 1-like (ENSG00000176092), score: 0.66 AKR1CL1aldo-keto reductase family 1, member C-like 1 (ENSG00000196326), score: 0.64 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: 0.64 ALOX12arachidonate 12-lipoxygenase (ENSG00000108839), score: 0.71 ANO7anoctamin 7 (ENSG00000146205), score: 0.57 ANXA10annexin A10 (ENSG00000109511), score: 0.52 APOA4apolipoprotein A-IV (ENSG00000110244), score: 0.66 AQP8aquaporin 8 (ENSG00000103375), score: 0.56 ARAP1ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 (ENSG00000186635), score: 0.52 ARSBarylsulfatase B (ENSG00000113273), score: 0.57 ART4ADP-ribosyltransferase 4 (Dombrock blood group) (ENSG00000111339), score: 0.61 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.51 B3GAT2beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) (ENSG00000112309), score: -0.49 B3GNT7UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 (ENSG00000156966), score: 0.66 BCAR3breast cancer anti-estrogen resistance 3 (ENSG00000137936), score: 0.51 BIKBCL2-interacting killer (apoptosis-inducing) (ENSG00000100290), score: 0.53 BIRC7baculoviral IAP repeat-containing 7 (ENSG00000101197), score: 0.61 C16orf91chromosome 16 open reading frame 91 (ENSG00000174109), score: 0.51 C17orf87chromosome 17 open reading frame 87 (ENSG00000161929), score: 0.53 C1orf177chromosome 1 open reading frame 177 (ENSG00000162398), score: 0.57 C20orf29chromosome 20 open reading frame 29 (ENSG00000125843), score: 0.51 C20orf7chromosome 20 open reading frame 7 (ENSG00000101247), score: -0.59 C22orf15chromosome 22 open reading frame 15 (ENSG00000169314), score: 0.56 C2orf43chromosome 2 open reading frame 43 (ENSG00000118961), score: 0.49 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.59 C3orf26chromosome 3 open reading frame 26 (ENSG00000184220), score: -0.55 C3orf37chromosome 3 open reading frame 37 (ENSG00000183624), score: -0.51 C9orf152chromosome 9 open reading frame 152 (ENSG00000188959), score: 0.74 CAMTA1calmodulin binding transcription activator 1 (ENSG00000171735), score: -0.56 CASP9caspase 9, apoptosis-related cysteine peptidase (ENSG00000132906), score: 0.53 CCDC142coiled-coil domain containing 142 (ENSG00000135637), score: 0.52 CCDC88Ccoiled-coil domain containing 88C (ENSG00000015133), score: 0.52 CCL23chemokine (C-C motif) ligand 23 (ENSG00000167236), score: 0.72 CCL24chemokine (C-C motif) ligand 24 (ENSG00000106178), score: 0.56 CCL25chemokine (C-C motif) ligand 25 (ENSG00000131142), score: 0.53 CD1ECD1e molecule (ENSG00000158488), score: 0.57 CD33CD33 molecule (ENSG00000105383), score: 0.51 CDC42BPACDC42 binding protein kinase alpha (DMPK-like) (ENSG00000143776), score: -0.6 CDX2caudal type homeobox 2 (ENSG00000165556), score: 0.6 CEACAM16carcinoembryonic antigen-related cell adhesion molecule 16 (ENSG00000213892), score: 0.5 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.56 CLDN14claudin 14 (ENSG00000159261), score: 0.52 CLDN15claudin 15 (ENSG00000106404), score: 0.52 CLINT1clathrin interactor 1 (ENSG00000113282), score: 0.53 CPA6carboxypeptidase A6 (ENSG00000165078), score: 0.5 CRYBA2crystallin, beta A2 (ENSG00000163499), score: 0.75 CXCR2chemokine (C-X-C motif) receptor 2 (ENSG00000180871), score: 0.59 CXCR3chemokine (C-X-C motif) receptor 3 (ENSG00000186810), score: 0.61 CYFIP1cytoplasmic FMR1 interacting protein 1 (ENSG00000068793), score: 0.54 CYP1A1cytochrome P450, family 1, subfamily A, polypeptide 1 (ENSG00000140465), score: 0.56 DEFB4Bdefensin, beta 4B (ENSG00000171711), score: 0.51 DKK2dickkopf homolog 2 (Xenopus laevis) (ENSG00000155011), score: 0.68 DNAJA4DnaJ (Hsp40) homolog, subfamily A, member 4 (ENSG00000140403), score: -0.53 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (ENSG00000168259), score: -0.51 DPYDdihydropyrimidine dehydrogenase (ENSG00000188641), score: 0.5 DSC3desmocollin 3 (ENSG00000134762), score: 0.75 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (ENSG00000134109), score: 0.49 EMR1egf-like module containing, mucin-like, hormone receptor-like 1 (ENSG00000174837), score: 0.57 ERLIN2ER lipid raft associated 2 (ENSG00000147475), score: 0.52 ESPL1extra spindle pole bodies homolog 1 (S. cerevisiae) (ENSG00000135476), score: 0.53 ESR1estrogen receptor 1 (ENSG00000091831), score: 0.49 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.75 FAM189Bfamily with sequence similarity 189, member B (ENSG00000160767), score: -0.64 FGD2FYVE, RhoGEF and PH domain containing 2 (ENSG00000146192), score: 0.52 FLJ35220hypothetical protein FLJ35220 (ENSG00000173818), score: -0.54 FOXP3forkhead box P3 (ENSG00000049768), score: 0.67 FPGSfolylpolyglutamate synthase (ENSG00000136877), score: 0.5 FPR3formyl peptide receptor 3 (ENSG00000187474), score: 0.55 FXC1fracture callus 1 homolog (rat) (ENSG00000132286), score: 0.51 FZD5frizzled homolog 5 (Drosophila) (ENSG00000163251), score: 0.5 GALNSgalactosamine (N-acetyl)-6-sulfate sulfatase (ENSG00000141012), score: 0.5 GAS8growth arrest-specific 8 (ENSG00000141013), score: 0.55 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.54 GLYATL3glycine-N-acyltransferase-like 3 (ENSG00000203972), score: 0.68 GNAT1guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 (ENSG00000114349), score: 0.65 GNAT2guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 (ENSG00000134183), score: 0.53 GPR152G protein-coupled receptor 152 (ENSG00000175514), score: 0.53 GPR172BG protein-coupled receptor 172B (ENSG00000132517), score: 0.5 GTSE1G-2 and S-phase expressed 1 (ENSG00000075218), score: 0.51 GUCA2Bguanylate cyclase activator 2B (uroguanylin) (ENSG00000044012), score: 0.51 GYLTL1Bglycosyltransferase-like 1B (ENSG00000165905), score: 0.55 HEATR3HEAT repeat containing 3 (ENSG00000155393), score: 0.49 HNRNPLheterogeneous nuclear ribonucleoprotein L (ENSG00000104824), score: -0.51 HRASLS2HRAS-like suppressor 2 (ENSG00000133328), score: 0.79 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (ENSG00000153976), score: 0.51 IFIT1Binterferon-induced protein with tetratricopeptide repeats 1B (ENSG00000204010), score: 0.55 IGFL3IGF-like family member 3 (ENSG00000188624), score: 0.59 IGSF6immunoglobulin superfamily, member 6 (ENSG00000140749), score: 0.52 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.51 IL2interleukin 2 (ENSG00000109471), score: 0.51 IVDisovaleryl-CoA dehydrogenase (ENSG00000128928), score: 0.57 IYDiodotyrosine deiodinase (ENSG00000009765), score: 0.49 JMJD4jumonji domain containing 4 (ENSG00000081692), score: 0.52 KIAA1609KIAA1609 (ENSG00000140950), score: 0.5 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.7 KRT28keratin 28 (ENSG00000173908), score: 0.85 LGMNlegumain (ENSG00000100600), score: 0.55 LIPMlipase, family member M (ENSG00000173239), score: 0.6 LOC100127905family with sequence similarity 165, member B pseudogene (ENSG00000198738), score: 0.55 LRRC8Eleucine rich repeat containing 8 family, member E (ENSG00000171017), score: 0.54 LUC7LLUC7-like (S. cerevisiae) (ENSG00000007392), score: -0.52 LYPD3LY6/PLAUR domain containing 3 (ENSG00000124466), score: 0.66 MCFD2multiple coagulation factor deficiency 2 (ENSG00000180398), score: 0.49 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.73 MPV17LMPV17 mitochondrial membrane protein-like (ENSG00000156968), score: 0.59 NGRNneugrin, neurite outgrowth associated (ENSG00000182768), score: -0.5 NSDHLNAD(P) dependent steroid dehydrogenase-like (ENSG00000147383), score: 0.52 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.57 NUMBnumb homolog (Drosophila) (ENSG00000133961), score: 0.56 NYXnyctalopin (ENSG00000188937), score: 0.58 OLA1Obg-like ATPase 1 (ENSG00000138430), score: -0.51 OPLAH5-oxoprolinase (ATP-hydrolysing) (ENSG00000178814), score: 0.49 OR10H3olfactory receptor, family 10, subfamily H, member 3 (ENSG00000171936), score: 0.66 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.57 OXToxytocin, prepropeptide (ENSG00000101405), score: 0.51 PAEPprogestagen-associated endometrial protein (ENSG00000122133), score: 0.67 PARP15poly (ADP-ribose) polymerase family, member 15 (ENSG00000173200), score: 0.54 PLA2G2Dphospholipase A2, group IID (ENSG00000117215), score: 0.69 PLEKHB2pleckstrin homology domain containing, family B (evectins) member 2 (ENSG00000115762), score: -0.5 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.6 PPAPDC1Bphosphatidic acid phosphatase type 2 domain containing 1B (ENSG00000147535), score: 0.64 PRHOXNBparahox cluster neighbor (ENSG00000183463), score: 0.83 PROL1proline rich, lacrimal 1 (ENSG00000171199), score: 0.57 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.7 PSKH2protein serine kinase H2 (ENSG00000147613), score: 0.73 RAB1BRAB1B, member RAS oncogene family (ENSG00000174903), score: -0.5 RANBP10RAN binding protein 10 (ENSG00000141084), score: 0.53 RBM6RNA binding motif protein 6 (ENSG00000004534), score: -0.5 RGS13regulator of G-protein signaling 13 (ENSG00000127074), score: 0.75 RHBDD3rhomboid domain containing 3 (ENSG00000100263), score: 0.52 RNASE1ribonuclease, RNase A family, 1 (pancreatic) (ENSG00000129538), score: -0.61 RPL36ALribosomal protein L36a-like (ENSG00000165502), score: -0.5 RPP40ribonuclease P/MRP 40kDa subunit (ENSG00000124787), score: 0.51 SCLT1sodium channel and clathrin linker 1 (ENSG00000151466), score: 0.53 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.56 SEC24ASEC24 family, member A (S. cerevisiae) (ENSG00000113615), score: 0.5 SERINC4serine incorporator 4 (ENSG00000184716), score: 0.61 SERPINA12serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 (ENSG00000165953), score: 0.69 SLC13A1solute carrier family 13 (sodium/sulfate symporters), member 1 (ENSG00000081800), score: 0.51 SLC15A5solute carrier family 15, member 5 (ENSG00000188991), score: 0.52 SLC16A11solute carrier family 16, member 11 (monocarboxylic acid transporter 11) (ENSG00000174326), score: 0.52 SLC19A3solute carrier family 19, member 3 (ENSG00000135917), score: 0.5 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.59 SLC23A1solute carrier family 23 (nucleobase transporters), member 1 (ENSG00000170482), score: 0.49 SLC45A3solute carrier family 45, member 3 (ENSG00000158715), score: 0.53 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.57 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (ENSG00000103064), score: -0.5 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.55 SMPDL3Asphingomyelin phosphodiesterase, acid-like 3A (ENSG00000172594), score: 0.57 SOAT2sterol O-acyltransferase 2 (ENSG00000167780), score: 0.59 SRD5A3steroid 5 alpha-reductase 3 (ENSG00000128039), score: 0.56 SRP14signal recognition particle 14kDa (homologous Alu RNA binding protein) (ENSG00000140319), score: -0.5 SYNCsyncoilin, intermediate filament protein (ENSG00000162520), score: -0.51 TBX19T-box 19 (ENSG00000143178), score: 0.64 TCEA2transcription elongation factor A (SII), 2 (ENSG00000171703), score: -0.56 TECTBtectorin beta (ENSG00000119913), score: 0.5 TLR6toll-like receptor 6 (ENSG00000174130), score: 0.67 TMEM61transmembrane protein 61 (ENSG00000143001), score: 0.5 TNFSF15tumor necrosis factor (ligand) superfamily, member 15 (ENSG00000181634), score: 0.53 TNNtenascin N (ENSG00000120332), score: 0.54 TRPM5transient receptor potential cation channel, subfamily M, member 5 (ENSG00000070985), score: 0.75 TRPM6transient receptor potential cation channel, subfamily M, member 6 (ENSG00000119121), score: 0.52 TTC21Btetratricopeptide repeat domain 21B (ENSG00000123607), score: -0.52 UCMAupper zone of growth plate and cartilage matrix associated (ENSG00000165623), score: 0.59 UCN3urocortin 3 (stresscopin) (ENSG00000178473), score: 0.5 VWCEvon Willebrand factor C and EGF domains (ENSG00000167992), score: 0.58 WDR55WD repeat domain 55 (ENSG00000120314), score: 0.51 XCR1chemokine (C motif) receptor 1 (ENSG00000173578), score: 0.49 XPCxeroderma pigmentosum, complementation group C (ENSG00000154767), score: 0.52 XYLBxylulokinase homolog (H. influenzae) (ENSG00000093217), score: 0.53 ZBTB3zinc finger and BTB domain containing 3 (ENSG00000185670), score: 0.53 ZNF226zinc finger protein 226 (ENSG00000167380), score: -0.5 ZNF496zinc finger protein 496 (ENSG00000162714), score: -0.49 ZNHIT3zinc finger, HIT type 3 (ENSG00000108278), score: -0.5 ZSWIM3zinc finger, SWIM-type containing 3 (ENSG00000132801), score: 0.5
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_kd_f_ca1 | mml | kd | f | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |