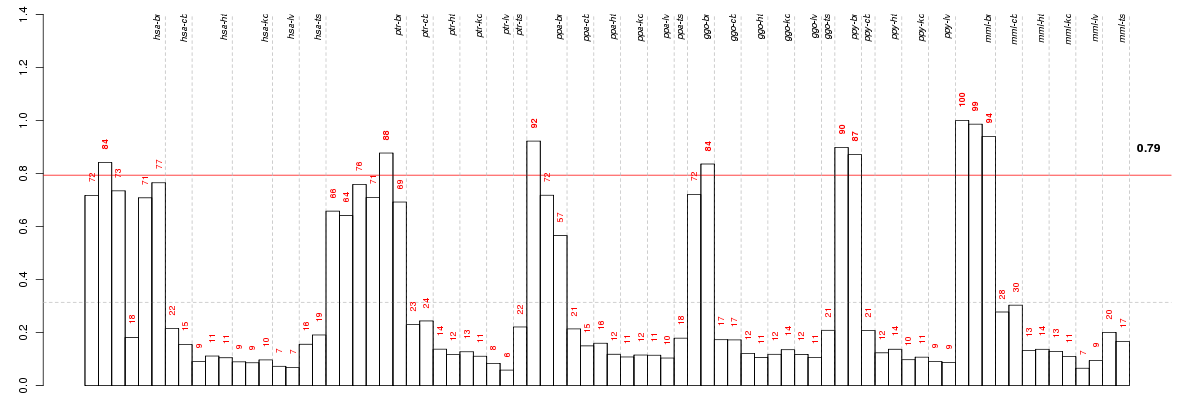

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cation transport
The directed movement of cations, atoms or small molecules with a net positive charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
potassium ion transport
The directed movement of potassium ions (K+) into, out of, within or between cells by means of some external agent such as a transporter or pore.
metal ion transport
The directed movement of metal ions, any metal ion with an electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
G-protein coupled receptor protein signaling pathway
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand.
G-protein signaling, coupled to cyclic nucleotide second messenger
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand, followed by modulation of a nucleotide cyclase activity and a subsequent change in the concentration of a cyclic nucleotide.
serotonin receptor signaling pathway
The series of molecular signals generated as a consequence of a serotonin receptor binding to one of its physiological ligands.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
behavior
The specific actions or reactions of an organism in response to external or internal stimuli. Patterned activity of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions.
learning or memory
The acquisition and processing of information and/or the storage and retrieval of this information over time.
memory
The activities involved in the mental information processing system that receives (registers), modifies, stores, and retrieves informational stimuli. The main stages involved in the formation and retrieval of memory are encoding (processing of received information by acquisition), storage (building a permanent record of received information as a result of consolidation) and retrieval (calling back the stored information and use it in a suitable way to execute a given task).
feeding behavior
Behavior associated with the intake of food.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
monovalent inorganic cation transport
The directed movement of inorganic cations with a valency of one into, out of, within or between cells by means of some external agent such as a transporter or pore. Inorganic cations are atoms or small molecules with a positive charge which do not contain carbon in covalent linkage.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
second-messenger-mediated signaling
A series of molecular signals in which an ion or small molecule is formed or released into the cytosol, thereby helping relay the signal within the cell.
cyclic-nucleotide-mediated signaling
A series of molecular signals in which a cell uses a cyclic nucleotide to convert an extracellular signal into a response.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
intracellular signaling pathway
The series of molecular events whereby information is sent from one location to another within a cell.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
regulation of system process
Any process that modulates the frequency, rate or extent of a system process, a multicellular organismal process carried out by any of the organs or tissues in an organ system.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cognition
The operation of the mind by which an organism becomes aware of objects of thought or perception; it includes the mental activities associated with thinking, learning, and memory.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
transmembrane transport
The process whereby a solute is transported from one side of a membrane to the other. This process includes the actual movement of the solute, and any regulation and preparatory steps, such as reduction of the solute.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
NA
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
regulation of system process
Any process that modulates the frequency, rate or extent of a system process, a multicellular organismal process carried out by any of the organs or tissues in an organ system.
transmembrane transport
The process whereby a solute is transported from one side of a membrane to the other. This process includes the actual movement of the solute, and any regulation and preparatory steps, such as reduction of the solute.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
learning or memory
The acquisition and processing of information and/or the storage and retrieval of this information over time.
potassium ion transport
The directed movement of potassium ions (K+) into, out of, within or between cells by means of some external agent such as a transporter or pore.
G-protein signaling, coupled to cyclic nucleotide second messenger
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand, followed by modulation of a nucleotide cyclase activity and a subsequent change in the concentration of a cyclic nucleotide.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
potassium channel complex
An ion channel complex through which potassium ions pass.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse
The junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell; the site of interneuronal communication. As the nerve fiber approaches the synapse it enlarges into a specialized structure, the presynaptic nerve ending, which contains mitochondria and synaptic vesicles. At the tip of the nerve ending is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic nerve ending secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane.
postsynaptic membrane
A specialized area of membrane facing the presynaptic membrane on the tip of the nerve ending and separated from it by a minute cleft (the synaptic cleft). Neurotransmitters across the synaptic cleft and transmit the signal to the postsynaptic membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
postsynaptic membrane
A specialized area of membrane facing the presynaptic membrane on the tip of the nerve ending and separated from it by a minute cleft (the synaptic cleft). Neurotransmitters across the synaptic cleft and transmit the signal to the postsynaptic membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.

peptide receptor activity
Combining with an extracellular or intracellular peptide to initiate a change in cell activity.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
inositol or phosphatidylinositol phosphodiesterase activity
NA
phosphoinositide phospholipase C activity
Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate + H2O = D-myo-inositol 1,4,5-trisphosphate + diacylglycerol.
phospholipase activity
Catalysis of the hydrolysis of a glycerophospholipid.
phospholipase C activity
Catalysis of the reaction: a phospholipid + H2O = 1,2-diacylglycerol + a phosphatidate.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
serotonin receptor activity
Combining with the biogenic amine serotonin, a neurotransmitter and hormone found in vertebrates, invertebrates and plants, to initiate a change in cell activity.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
voltage-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons.
voltage-gated potassium channel activity
Catalysis of the transmembrane transfer of a potassium ion by a voltage-gated channel.
delayed rectifier potassium channel activity
Catalysis of the transmembrane transfer of a potassium ion by a delayed rectifying voltage-gated channel. A delayed rectifying current-voltage relation is one where channel activation kinetics are time-dependent, and activation is slow.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
potassium channel activity
Catalysis of facilitated diffusion of a potassium ion (by an energy-independent process) involving passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
phosphoric diester hydrolase activity
Catalysis of the hydrolysis of a phosphodiester to give a phosphomonoester and a free hydroxyl group.
drug binding
Interacting selectively and non-covalently with a drug, any naturally occurring or synthetic substance, other than a nutrient, that, when administered or applied to an organism, affects the structure or functioning of the organism; in particular, any such substance used in the diagnosis, prevention, or treatment of disease.
G-protein coupled amine receptor activity
A receptor that binds an extracellular amine and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
peptide receptor activity, G-protein coupled
Combining with an extracellular or intracellular peptide to initiate a G-protein mediated change in cell activity. A G-protein is a signal transduction molecule that alternates between an inactive GDP-bound and an active GTP-bound state.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
lipase activity
Catalysis of the hydrolysis of a lipid or phospholipid.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on ester bonds
Catalysis of the hydrolysis of any ester bond.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
voltage-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel whose open state is dependent on the voltage across the membrane in which it is embedded.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
voltage-gated cation channel activity
Catalysis of the transmembrane transfer of a cation by a voltage-gated channel. A cation is a positively charged ion.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
neurotransmitter binding
Interacting selectively and non-covalently with a neurotransmitter, any chemical substance that is capable of transmitting (or inhibiting the transmission of) a nerve impulse from a neuron to another cell.
peptide binding
Interacting selectively and non-covalently with peptides, any of a group of organic compounds comprising two or more amino acids linked by peptide bonds.
phosphoric ester hydrolase activity
Catalysis of the reaction: RPO-R' + H2O = RPOOH + R'H. This reaction is the hydrolysis of any phosphoric ester bond, any ester formed from orthophosphoric acid, O=P(OH)3.
amine binding
Interacting selectively and non-covalently with any organic compound that is weakly basic in character and contains an amino or a substituted amino group.
serotonin binding
Interacting selectively and non-covalently with serotonin (5-hydroxytryptamine), a monoamine neurotransmitter occurring in the peripheral and central nervous systems, also having hormonal properties.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
peptide receptor activity
Combining with an extracellular or intracellular peptide to initiate a change in cell activity.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
phospholipase C activity
Catalysis of the reaction: a phospholipid + H2O = 1,2-diacylglycerol + a phosphatidate.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
peptide receptor activity, G-protein coupled
Combining with an extracellular or intracellular peptide to initiate a G-protein mediated change in cell activity. A G-protein is a signal transduction molecule that alternates between an inactive GDP-bound and an active GTP-bound state.
phosphoinositide phospholipase C activity
Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate + H2O = D-myo-inositol 1,4,5-trisphosphate + diacylglycerol.
voltage-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons.
voltage-gated cation channel activity
Catalysis of the transmembrane transfer of a cation by a voltage-gated channel. A cation is a positively charged ion.
voltage-gated potassium channel activity
Catalysis of the transmembrane transfer of a potassium ion by a voltage-gated channel.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 3.839e-10 | 2.094 | 17 | 206 | Neuroactive ligand-receptor interaction |
| 04020 | 4.165e-04 | 1.474 | 9 | 145 | Calcium signaling pathway |
| 04540 | 7.116e-03 | 0.5998 | 5 | 59 | Gap junction |
ANO3anoctamin 3 (ENSG00000134343), score: 0.84 ATRNL1attractin-like 1 (ENSG00000107518), score: 0.78 B3GALT1UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 (ENSG00000172318), score: 0.82 BTBD9BTB (POZ) domain containing 9 (ENSG00000183826), score: 0.78 C13orf36chromosome 13 open reading frame 36 (ENSG00000180440), score: 0.87 CACNG3calcium channel, voltage-dependent, gamma subunit 3 (ENSG00000006116), score: 0.8 CBLN4cerebellin 4 precursor (ENSG00000054803), score: 0.85 CDH12cadherin 12, type 2 (N-cadherin 2) (ENSG00000154162), score: 0.85 CDH8cadherin 8, type 2 (ENSG00000150394), score: 0.91 CDH9cadherin 9, type 2 (T1-cadherin) (ENSG00000113100), score: 0.87 CDKL5cyclin-dependent kinase-like 5 (ENSG00000008086), score: 0.77 CHRM3cholinergic receptor, muscarinic 3 (ENSG00000133019), score: 0.86 CNTN3contactin 3 (plasmacytoma associated) (ENSG00000113805), score: 0.9 CNTN5contactin 5 (ENSG00000149972), score: 0.81 CNTNAP5contactin associated protein-like 5 (ENSG00000155052), score: 0.88 CRHcorticotropin releasing hormone (ENSG00000147571), score: 0.84 CSMD3CUB and Sushi multiple domains 3 (ENSG00000164796), score: 0.86 CSRNP3cysteine-serine-rich nuclear protein 3 (ENSG00000178662), score: 0.92 DCXdoublecortin (ENSG00000077279), score: 0.79 DGKBdiacylglycerol kinase, beta 90kDa (ENSG00000136267), score: 0.85 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.81 DLX1distal-less homeobox 1 (ENSG00000144355), score: 0.81 DPP10dipeptidyl-peptidase 10 (non-functional) (ENSG00000175497), score: 0.79 ENC1ectodermal-neural cortex 1 (with BTB-like domain) (ENSG00000171617), score: 0.77 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.85 FAM19A1family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 (ENSG00000183662), score: 0.86 FAM19A2family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 (ENSG00000198673), score: 0.85 FAM49Afamily with sequence similarity 49, member A (ENSG00000197872), score: 0.78 FAR2fatty acyl CoA reductase 2 (ENSG00000064763), score: 0.78 FERD3LFer3-like (Drosophila) (ENSG00000146618), score: 0.89 FGF10fibroblast growth factor 10 (ENSG00000070193), score: 0.93 FLRT2fibronectin leucine rich transmembrane protein 2 (ENSG00000185070), score: 0.79 FRMPD4FERM and PDZ domain containing 4 (ENSG00000169933), score: 0.78 GABRA4gamma-aminobutyric acid (GABA) A receptor, alpha 4 (ENSG00000109158), score: 0.92 GALNTL6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 (ENSG00000174473), score: 0.84 GJD2gap junction protein, delta 2, 36kDa (ENSG00000159248), score: 0.8 GLRA3glycine receptor, alpha 3 (ENSG00000145451), score: 1 GPR149G protein-coupled receptor 149 (ENSG00000174948), score: 0.77 GPR26G protein-coupled receptor 26 (ENSG00000154478), score: 0.85 GPR83G protein-coupled receptor 83 (ENSG00000123901), score: 0.76 GRIN2Bglutamate receptor, ionotropic, N-methyl D-aspartate 2B (ENSG00000150086), score: 0.82 HRH1histamine receptor H1 (ENSG00000196639), score: 0.83 HTR1A5-hydroxytryptamine (serotonin) receptor 1A (ENSG00000178394), score: 0.8 HTR1B5-hydroxytryptamine (serotonin) receptor 1B (ENSG00000135312), score: 0.79 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (ENSG00000102468), score: 0.86 HTR2C5-hydroxytryptamine (serotonin) receptor 2C (ENSG00000147246), score: 0.85 HTR45-hydroxytryptamine (serotonin) receptor 4 (ENSG00000164270), score: 0.85 HTR65-hydroxytryptamine (serotonin) receptor 6 (ENSG00000158748), score: 0.87 KAL1Kallmann syndrome 1 sequence (ENSG00000011201), score: 0.79 KCNA3potassium voltage-gated channel, shaker-related subfamily, member 3 (ENSG00000177272), score: 0.77 KCNA4potassium voltage-gated channel, shaker-related subfamily, member 4 (ENSG00000182255), score: 0.86 KCNB2potassium voltage-gated channel, Shab-related subfamily, member 2 (ENSG00000182674), score: 0.88 KCNC2potassium voltage-gated channel, Shaw-related subfamily, member 2 (ENSG00000166006), score: 0.79 KCNH4potassium voltage-gated channel, subfamily H (eag-related), member 4 (ENSG00000089558), score: 0.82 KCNH7potassium voltage-gated channel, subfamily H (eag-related), member 7 (ENSG00000184611), score: 0.91 KCNMB2potassium large conductance calcium-activated channel, subfamily M, beta member 2 (ENSG00000197584), score: 0.8 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (ENSG00000184156), score: 0.77 KCNQ5potassium voltage-gated channel, KQT-like subfamily, member 5 (ENSG00000185760), score: 0.82 KCNS1potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 (ENSG00000124134), score: 0.82 KCNS2potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 (ENSG00000156486), score: 0.86 KCNV1potassium channel, subfamily V, member 1 (ENSG00000164794), score: 0.83 KCTD16potassium channel tetramerisation domain containing 16 (ENSG00000183775), score: 0.83 KCTD4potassium channel tetramerisation domain containing 4 (ENSG00000180332), score: 0.8 KIAA0748KIAA0748 (ENSG00000135426), score: 0.78 KIAA2022KIAA2022 (ENSG00000050030), score: 0.83 LOC100293905similar to gastrin-releasing peptide receptor (ENSG00000126010), score: 0.94 LPPR4lipid phosphate phosphatase-related protein type 4 (ENSG00000117600), score: 0.78 LPPR5lipid phosphate phosphatase-related protein type 5 (ENSG00000117598), score: 0.82 LRFN5leucine rich repeat and fibronectin type III domain containing 5 (ENSG00000165379), score: 0.77 LRRC55leucine rich repeat containing 55 (ENSG00000183908), score: 0.82 MAPK8mitogen-activated protein kinase 8 (ENSG00000107643), score: 0.76 MAS1MAS1 oncogene (ENSG00000130368), score: 0.92 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.84 MEF2Cmyocyte enhancer factor 2C (ENSG00000081189), score: 0.76 MEPEmatrix extracellular phosphoglycoprotein (ENSG00000152595), score: 0.83 MKL2MKL/myocardin-like 2 (ENSG00000186260), score: 0.81 MMP16matrix metallopeptidase 16 (membrane-inserted) (ENSG00000156103), score: 0.78 NCALDneurocalcin delta (ENSG00000104490), score: 0.77 NECAB1N-terminal EF-hand calcium binding protein 1 (ENSG00000123119), score: 0.81 NETO1neuropilin (NRP) and tolloid (TLL)-like 1 (ENSG00000166342), score: 0.87 NEUROD6neurogenic differentiation 6 (ENSG00000164600), score: 0.79 NLKnemo-like kinase (ENSG00000087095), score: 0.83 NPEPL1aminopeptidase-like 1 (ENSG00000215440), score: -0.79 NPY2Rneuropeptide Y receptor Y2 (ENSG00000185149), score: 0.88 NRG3neuregulin 3 (ENSG00000185737), score: 0.8 OPRD1opioid receptor, delta 1 (ENSG00000116329), score: 0.82 OSTNosteocrin (ENSG00000188729), score: 0.83 PCDH19protocadherin 19 (ENSG00000165194), score: 0.85 PCDH20protocadherin 20 (ENSG00000197991), score: 0.82 PCSK1proprotein convertase subtilisin/kexin type 1 (ENSG00000175426), score: 0.87 PDYNprodynorphin (ENSG00000101327), score: 0.87 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (ENSG00000182621), score: 0.82 PNOCprepronociceptin (ENSG00000168081), score: 0.78 PRKG2protein kinase, cGMP-dependent, type II (ENSG00000138669), score: 0.98 PRRG3proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) (ENSG00000130032), score: 0.78 RGS4regulator of G-protein signaling 4 (ENSG00000117152), score: 0.81 RORBRAR-related orphan receptor B (ENSG00000198963), score: 0.83 RSPO2R-spondin 2 homolog (Xenopus laevis) (ENSG00000147655), score: 0.77 RXFP3relaxin/insulin-like family peptide receptor 3 (ENSG00000182631), score: 0.95 SATB2SATB homeobox 2 (ENSG00000119042), score: 0.76 SCN3Asodium channel, voltage-gated, type III, alpha subunit (ENSG00000153253), score: 0.76 SEMA3Asema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A (ENSG00000075213), score: 0.84 SGTBsmall glutamine-rich tetratricopeptide repeat (TPR)-containing, beta (ENSG00000197860), score: 0.78 SHC3SHC (Src homology 2 domain containing) transforming protein 3 (ENSG00000148082), score: 0.82 SLC24A4solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 (ENSG00000140090), score: 0.77 SLC39A10solute carrier family 39 (zinc transporter), member 10 (ENSG00000196950), score: 0.78 SNTG1syntrophin, gamma 1 (ENSG00000147481), score: 0.78 SPIN1spindlin 1 (ENSG00000106723), score: 0.79 ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (ENSG00000117069), score: 0.8 STYK1serine/threonine/tyrosine kinase 1 (ENSG00000060140), score: 0.86 TBR1T-box, brain, 1 (ENSG00000136535), score: 0.76 TMEM132Dtransmembrane protein 132D (ENSG00000151952), score: 0.85 TMEM155transmembrane protein 155 (ENSG00000164112), score: 0.81 TMEM196transmembrane protein 196 (ENSG00000173452), score: 0.85 TMEM200Atransmembrane protein 200A (ENSG00000164484), score: 0.76 TRHRthyrotropin-releasing hormone receptor (ENSG00000174417), score: 0.91 TRPC5transient receptor potential cation channel, subfamily C, member 5 (ENSG00000072315), score: 0.87 UNC5Dunc-5 homolog D (C. elegans) (ENSG00000156687), score: 0.8 VIPvasoactive intestinal peptide (ENSG00000146469), score: 0.87 VSTM2AV-set and transmembrane domain containing 2A (ENSG00000170419), score: 0.83 VWC2Lvon Willebrand factor C domain-containing protein 2-like (ENSG00000174453), score: 0.94 XKR4XK, Kell blood group complex subunit-related family, member 4 (ENSG00000206579), score: 0.77 ZNF804Bzinc finger protein 804B (ENSG00000182348), score: 0.78 ZNF831zinc finger protein 831 (ENSG00000124203), score: 0.82
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_br_f_ca1 | ggo | br | f | _ |
| hsa_br_m2_ca1 | hsa | br | m | 2 |
| ppy_br_f_ca1 | ppy | br | f | _ |
| ptr_br_m4_ca1 | ptr | br | m | 4 |
| ppy_br_m_ca1 | ppy | br | m | _ |
| ppa_br_m_ca1 | ppa | br | m | _ |
| mml_br_f_ca1 | mml | br | f | _ |
| mml_br_m1_ca1 | mml | br | m | 1 |
| mml_br_m2_ca1 | mml | br | m | 2 |