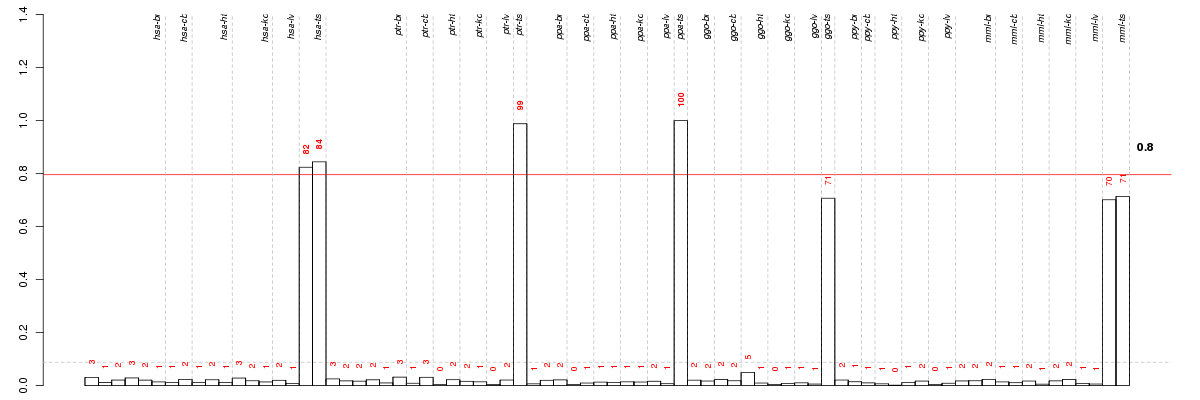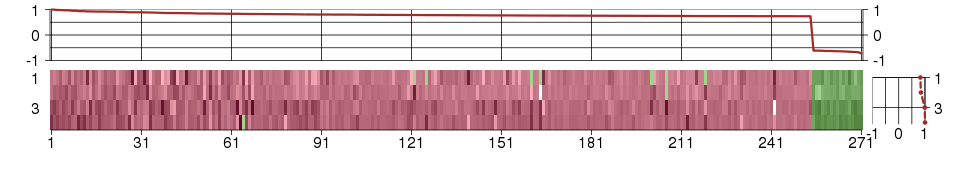

Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
reproductive developmental process
A developmental process by which a progressive change in the state of some part of an organism specifically contributes to its ability to form offspring.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The cellular synthesis of RNA on a template of DNA.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
gamete generation
The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell.
spermatogenesis
The process of formation of spermatozoa, including spermatocytogenesis and spermiogenesis.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
sexual reproduction
The regular alternation, in the life cycle of haplontic, diplontic and diplohaplontic organisms, of meiosis and fertilization which provides for the production offspring. In diplontic organisms there is a life cycle in which the products of meiosis behave directly as gametes, fusing to form a zygote from which the diploid, or sexually reproductive polyploid, adult organism will develop. In diplohaplontic organisms a haploid phase (gametophyte) exists in the life cycle between meiosis and fertilization (e.g. higher plants, many algae and Fungi); the products of meiosis are spores that develop as haploid individuals from which haploid gametes develop to form a diploid zygote; diplohaplontic organisms show an alternation of haploid and diploid generations. In haplontic organisms meiosis occurs in the zygote, giving rise to four haploid cells (e.g. many algae and protozoa), only the zygote is diploid and this may form a resistant spore, tiding organisms over hard times.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
male gamete generation
Generation of the male gamete; specialised haploid cells produced by meiosis and along with a female gamete takes part in sexual reproduction.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
reproductive cellular process
A process, occurring at the cellular level, that is involved in the reproductive function of a multicellular or single-celled organism.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
reproductive cellular process
A process, occurring at the cellular level, that is involved in the reproductive function of a multicellular or single-celled organism.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
reproductive developmental process
A developmental process by which a progressive change in the state of some part of an organism specifically contributes to its ability to form offspring.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
gamete generation
The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The cellular synthesis of RNA on a template of DNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
acrosomal vesicle
A structure in the head of a spermatozoon that contains acid hydrolases, and is concerned with the breakdown of the outer membrane of the ovum during fertilization. It lies just beneath the plasma membrane and is derived from the lysosome.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
stored secretory granule
A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell and upon stimulation, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
DNA binding
Any molecular function by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
sequence-specific DNA binding transcription factor activity
Interacting selectively and non-covalently with a specific DNA sequence in order to modulate transcription. The transcription factor may or may not also interact selectively with a protein or macromolecular complex.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
sequence-specific DNA binding
Interacting selectively and non-covalently with DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding.
all
NA
AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.86 ABLIM3actin binding LIM protein family, member 3 (ENSG00000173210), score: -0.63 ACRacrosin (ENSG00000100312), score: 0.79 ACTL7Aactin-like 7A (ENSG00000187003), score: 0.76 ACTL8actin-like 8 (ENSG00000117148), score: 0.85 ACTRT2actin-related protein T2 (ENSG00000169717), score: 0.74 ADAD1adenosine deaminase domain containing 1 (testis-specific) (ENSG00000164113), score: 0.75 ADAD2adenosine deaminase domain containing 2 (ENSG00000140955), score: 0.77 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (ENSG00000129221), score: 0.92 ANKFN1ankyrin-repeat and fibronectin type III domain containing 1 (ENSG00000153930), score: 0.77 ANKFY1ankyrin repeat and FYVE domain containing 1 (ENSG00000185722), score: -0.61 ANP32Aacidic (leucine-rich) nuclear phosphoprotein 32 family, member A (ENSG00000140350), score: -0.63 AQP10aquaporin 10 (ENSG00000143595), score: 0.77 AQP5aquaporin 5 (ENSG00000161798), score: 0.79 ARMC3armadillo repeat containing 3 (ENSG00000165309), score: 0.75 ARMC4armadillo repeat containing 4 (ENSG00000169126), score: 0.74 ATP6V1E2ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 (ENSG00000171142), score: 0.74 AURKCaurora kinase C (ENSG00000105146), score: 0.8 BARX1BARX homeobox 1 (ENSG00000131668), score: 0.76 BMP15bone morphogenetic protein 15 (ENSG00000130385), score: 0.95 BOLLbol, boule-like (Drosophila) (ENSG00000152430), score: 0.75 BTG4B-cell translocation gene 4 (ENSG00000137707), score: 0.76 BUB1budding uninhibited by benzimidazoles 1 homolog (yeast) (ENSG00000169679), score: 0.76 C11orf88chromosome 11 open reading frame 88 (ENSG00000183644), score: 0.79 C12orf12chromosome 12 open reading frame 12 (ENSG00000197651), score: 0.78 C13orf28chromosome 13 open reading frame 28 (ENSG00000153498), score: 0.75 C15orf39chromosome 15 open reading frame 39 (ENSG00000167173), score: 0.74 C15orf55chromosome 15 open reading frame 55 (ENSG00000184507), score: 0.79 C15orf60chromosome 15 open reading frame 60 (ENSG00000183324), score: 0.78 C17orf50chromosome 17 open reading frame 50 (ENSG00000154768), score: 0.85 C17orf53chromosome 17 open reading frame 53 (ENSG00000125319), score: 0.75 C17orf66chromosome 17 open reading frame 66 (ENSG00000172653), score: 0.8 C17orf74chromosome 17 open reading frame 74 (ENSG00000184560), score: 0.78 C17orf98chromosome 17 open reading frame 98 (ENSG00000214556), score: 0.74 C19orf45chromosome 19 open reading frame 45 (ENSG00000198723), score: 0.8 C1orf94chromosome 1 open reading frame 94 (ENSG00000142698), score: 0.74 C20orf151chromosome 20 open reading frame 151 (ENSG00000130701), score: 0.81 C20orf195chromosome 20 open reading frame 195 (ENSG00000125531), score: 0.74 C20orf71chromosome 20 open reading frame 71 (ENSG00000131059), score: 0.77 C20orf85chromosome 20 open reading frame 85 (ENSG00000124237), score: 0.79 C22orf23chromosome 22 open reading frame 23 (ENSG00000128346), score: 0.76 C2orf65chromosome 2 open reading frame 65 (ENSG00000159374), score: 0.74 C2orf78chromosome 2 open reading frame 78 (ENSG00000187833), score: 0.75 C3orf20chromosome 3 open reading frame 20 (ENSG00000131379), score: 0.74 C3orf30chromosome 3 open reading frame 30 (ENSG00000163424), score: 0.75 C4orf40chromosome 4 open reading frame 40 (ENSG00000187533), score: 0.83 C5orf47chromosome 5 open reading frame 47 (ENSG00000185056), score: 0.82 C5orf52chromosome 5 open reading frame 52 (ENSG00000187658), score: 0.78 CAPN1calpain 1, (mu/I) large subunit (ENSG00000014216), score: -0.67 CASC5cancer susceptibility candidate 5 (ENSG00000137812), score: 0.76 CATSPER4cation channel, sperm associated 4 (ENSG00000188782), score: 0.81 CCDC105coiled-coil domain containing 105 (ENSG00000160994), score: 0.79 CCDC27coiled-coil domain containing 27 (ENSG00000162592), score: 0.76 CCDC86coiled-coil domain containing 86 (ENSG00000110104), score: 0.74 CCDC96coiled-coil domain containing 96 (ENSG00000173013), score: 0.78 CDC20Bcell division cycle 20 homolog B (S. cerevisiae) (ENSG00000164287), score: 0.81 CDC25Ccell division cycle 25 homolog C (S. pombe) (ENSG00000158402), score: 0.76 CDC45cell division cycle 45 homolog (S. cerevisiae) (ENSG00000093009), score: 0.81 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.8 CETN1centrin, EF-hand protein, 1 (ENSG00000177143), score: 0.74 CHATcholine O-acetyltransferase (ENSG00000070748), score: 0.77 CLSPNclaspin (ENSG00000092853), score: 0.78 CNGA4cyclic nucleotide gated channel alpha 4 (ENSG00000132259), score: 0.76 COL17A1collagen, type XVII, alpha 1 (ENSG00000065618), score: 0.74 COL2A1collagen, type II, alpha 1 (ENSG00000139219), score: 0.8 COMMD7COMM domain containing 7 (ENSG00000149600), score: -0.65 CPA5carboxypeptidase A5 (ENSG00000158525), score: 0.78 CRYGCcrystallin, gamma C (ENSG00000163254), score: 0.76 CSN3casein kappa (ENSG00000171209), score: 0.75 CSNK2A2casein kinase 2, alpha prime polypeptide (ENSG00000070770), score: 0.82 CTCFLCCCTC-binding factor (zinc finger protein)-like (ENSG00000124092), score: 0.79 CXorf67chromosome X open reading frame 67 (ENSG00000187690), score: 0.75 DEFB119defensin, beta 119 (ENSG00000180483), score: 0.78 DEFB121defensin, beta 121 (ENSG00000204548), score: 0.89 DKFZP781G0119hypothetical protein LOC644041 (ENSG00000206043), score: 0.76 DKK4dickkopf homolog 4 (Xenopus laevis) (ENSG00000104371), score: 0.84 DMBX1diencephalon/mesencephalon homeobox 1 (ENSG00000197587), score: 0.92 DMRT3doublesex and mab-3 related transcription factor 3 (ENSG00000064218), score: 0.91 DMRTB1DMRT-like family B with proline-rich C-terminal, 1 (ENSG00000143006), score: 0.82 DMRTC2DMRT-like family C2 (ENSG00000142025), score: 0.8 DNAI2dynein, axonemal, intermediate chain 2 (ENSG00000171595), score: 0.75 DNAJB8DnaJ (Hsp40) homolog, subfamily B, member 8 (ENSG00000179407), score: 0.74 DNAJC5BDnaJ (Hsp40) homolog, subfamily C, member 5 beta (ENSG00000147570), score: 0.74 DPEP3dipeptidase 3 (ENSG00000141096), score: 0.76 DRG1developmentally regulated GTP binding protein 1 (ENSG00000185721), score: 0.84 DSG4desmoglein 4 (ENSG00000175065), score: 0.82 ESX1ESX homeobox 1 (ENSG00000123576), score: 0.84 EVX1even-skipped homeobox 1 (ENSG00000106038), score: 0.94 FAM154Afamily with sequence similarity 154, member A (ENSG00000155875), score: 0.76 FAM164Cfamily with sequence similarity 164, member C (ENSG00000119703), score: 0.75 FAM177Bfamily with sequence similarity 177, member B (ENSG00000197520), score: 0.76 FAM47Bfamily with sequence similarity 47, member B (ENSG00000189132), score: 0.83 FAM47Cfamily with sequence similarity 47, member C (ENSG00000198173), score: 0.8 FAM55Afamily with sequence similarity 55, member A (ENSG00000095110), score: 0.97 FAM71F1family with sequence similarity 71, member F1 (ENSG00000135248), score: 0.74 FAM9Cfamily with sequence similarity 9, member C (ENSG00000187268), score: 0.77 FATE1fetal and adult testis expressed 1 (ENSG00000147378), score: 0.74 FER1L6fer-1-like 6 (C. elegans) (ENSG00000214814), score: 0.75 FGF4fibroblast growth factor 4 (ENSG00000075388), score: 1 FOXN4forkhead box N4 (ENSG00000139445), score: 0.85 FOXR1forkhead box R1 (ENSG00000176302), score: 0.8 FSCN3fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus) (ENSG00000106328), score: 0.75 FTMTferritin mitochondrial (ENSG00000181867), score: 0.75 G6PC2glucose-6-phosphatase, catalytic, 2 (ENSG00000152254), score: 0.99 GAB4GRB2-associated binding protein family, member 4 (ENSG00000215568), score: 0.92 GBX1gastrulation brain homeobox 1 (ENSG00000164900), score: 0.77 GGA1golgi-associated, gamma adaptin ear containing, ARF binding protein 1 (ENSG00000100083), score: 0.78 GJA8gap junction protein, alpha 8, 50kDa (ENSG00000121634), score: 0.91 GLRA4glycine receptor, alpha 4 (ENSG00000188828), score: 0.8 GPR156G protein-coupled receptor 156 (ENSG00000175697), score: 0.79 GTSF1gametocyte specific factor 1 (ENSG00000170627), score: 0.76 GUCY2Dguanylate cyclase 2D, membrane (retina-specific) (ENSG00000132518), score: 0.77 HDAC8histone deacetylase 8 (ENSG00000147099), score: -0.72 HDGFL1hepatoma derived growth factor-like 1 (ENSG00000112273), score: 0.74 HERC3hect domain and RLD 3 (ENSG00000138641), score: -0.62 HOXB13homeobox B13 (ENSG00000159184), score: 0.78 HOXC13homeobox C13 (ENSG00000123364), score: 0.83 HSF5heat shock transcription factor family member 5 (ENSG00000176160), score: 0.74 HTR75-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled) (ENSG00000148680), score: 0.74 IL13interleukin 13 (ENSG00000169194), score: 0.88 IL23Rinterleukin 23 receptor (ENSG00000162594), score: 0.77 IL3interleukin 3 (colony-stimulating factor, multiple) (ENSG00000164399), score: 0.89 INSL6insulin-like 6 (ENSG00000120210), score: 0.77 KCNV2potassium channel, subfamily V, member 2 (ENSG00000168263), score: 0.97 KDM4Alysine (K)-specific demethylase 4A (ENSG00000066135), score: -0.63 KELKell blood group, metallo-endopeptidase (ENSG00000197993), score: 0.81 KIF24kinesin family member 24 (ENSG00000186638), score: 0.77 KLF17Kruppel-like factor 17 (ENSG00000171872), score: 0.79 KLF9Kruppel-like factor 9 (ENSG00000119138), score: -0.62 KLHL10kelch-like 10 (Drosophila) (ENSG00000161594), score: 0.74 KLK12kallikrein-related peptidase 12 (ENSG00000186474), score: 0.93 KLK13kallikrein-related peptidase 13 (ENSG00000167759), score: 0.97 KRT1keratin 1 (ENSG00000167768), score: 0.76 KRT26keratin 26 (ENSG00000186393), score: 0.89 LALBAlactalbumin, alpha- (ENSG00000167531), score: 0.85 LAMP3lysosomal-associated membrane protein 3 (ENSG00000078081), score: 0.98 LCTLlactase-like (ENSG00000188501), score: 0.74 LDHAL6Blactate dehydrogenase A-like 6B (ENSG00000171989), score: 0.74 LEMD1LEM domain containing 1 (ENSG00000186007), score: 0.74 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.98 LIM2lens intrinsic membrane protein 2, 19kDa (ENSG00000105370), score: 0.89 LIN28Alin-28 homolog A (C. elegans) (ENSG00000131914), score: 0.8 LOC100287738hypothetical protein LOC100287738 (ENSG00000139656), score: 0.84 LOC100292575similar to melanoma antigen family C, 2 (ENSG00000046774), score: 0.9 LOC388946transmembrane protein ENSP00000343375 (ENSG00000242691), score: 0.75 LOXHD1lipoxygenase homology domains 1 (ENSG00000167210), score: 0.86 LRIT1leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 (ENSG00000148602), score: 0.94 LRRC18leucine rich repeat containing 18 (ENSG00000165383), score: 0.79 LUZP4leucine zipper protein 4 (ENSG00000102021), score: 0.82 LYPD4LY6/PLAUR domain containing 4 (ENSG00000183103), score: 0.76 LYZL6lysozyme-like 6 (ENSG00000161572), score: 0.74 MAGEA1melanoma antigen family A, 1 (directs expression of antigen MZ2-E) (ENSG00000198681), score: 0.89 MAGEA11melanoma antigen family A, 11 (ENSG00000185247), score: 0.92 MAGEA4melanoma antigen family A, 4 (ENSG00000147381), score: 0.82 MAGEB2melanoma antigen family B, 2 (ENSG00000099399), score: 0.84 MBD1methyl-CpG binding domain protein 1 (ENSG00000141644), score: 0.78 MBD3L1methyl-CpG binding domain protein 3-like 1 (ENSG00000170948), score: 0.75 MEP1Ameprin A, alpha (PABA peptide hydrolase) (ENSG00000112818), score: 0.88 MFSD6Lmajor facilitator superfamily domain containing 6-like (ENSG00000185156), score: 0.76 MGLLmonoglyceride lipase (ENSG00000074416), score: -0.63 MMP20matrix metallopeptidase 20 (ENSG00000137674), score: 0.86 MS4A6Emembrane-spanning 4-domains, subfamily A, member 6E (ENSG00000166926), score: 0.91 NANOS2nanos homolog 2 (Drosophila) (ENSG00000188425), score: 0.88 NKX2-4NK2 homeobox 4 (ENSG00000125816), score: 0.87 NKX6-1NK6 homeobox 1 (ENSG00000163623), score: 0.76 NLRP7NLR family, pyrin domain containing 7 (ENSG00000167634), score: 0.91 NR0B1nuclear receptor subfamily 0, group B, member 1 (ENSG00000169297), score: 0.77 NR6A1nuclear receptor subfamily 6, group A, member 1 (ENSG00000148200), score: 0.74 NUCKS1nuclear casein kinase and cyclin-dependent kinase substrate 1 (ENSG00000069275), score: -0.66 NUP210Lnucleoporin 210kDa-like (ENSG00000143552), score: 0.76 NUP98nucleoporin 98kDa (ENSG00000110713), score: 0.77 ODF3L1outer dense fiber of sperm tails 3-like 1 (ENSG00000182950), score: 0.78 OOEPoocyte expressed protein homolog (dog) (ENSG00000203907), score: 0.84 OPN5opsin 5 (ENSG00000124818), score: 0.95 OR4N2olfactory receptor, family 4, subfamily N, member 2 (ENSG00000176294), score: 0.78 OR56A1olfactory receptor, family 56, subfamily A, member 1 (ENSG00000180934), score: 0.74 OR6F1olfactory receptor, family 6, subfamily F, member 1 (ENSG00000169214), score: 0.84 OTPorthopedia homeobox (ENSG00000171540), score: 0.74 PADI1peptidyl arginine deiminase, type I (ENSG00000142623), score: 0.84 PAGE1P antigen family, member 1 (prostate associated) (ENSG00000068985), score: 0.83 PASD1PAS domain containing 1 (ENSG00000166049), score: 0.89 PAX4paired box 4 (ENSG00000106331), score: 0.84 PBKPDZ binding kinase (ENSG00000168078), score: 0.75 PDE6Aphosphodiesterase 6A, cGMP-specific, rod, alpha (ENSG00000132915), score: 0.91 PDHA2pyruvate dehydrogenase (lipoamide) alpha 2 (ENSG00000163114), score: 0.77 PDZD11PDZ domain containing 11 (ENSG00000120509), score: -0.66 PFN4profilin family, member 4 (ENSG00000176732), score: 0.77 PHOX2Apaired-like homeobox 2a (ENSG00000165462), score: 0.86 PITX1paired-like homeodomain 1 (ENSG00000069011), score: 0.86 PIWIL1piwi-like 1 (Drosophila) (ENSG00000125207), score: 0.74 PIWIL2piwi-like 2 (Drosophila) (ENSG00000197181), score: 0.74 PLA2G2Ephospholipase A2, group IIE (ENSG00000188784), score: 0.75 POM121L2POM121 membrane glycoprotein-like 2 (ENSG00000158553), score: 0.82 POU4F2POU class 4 homeobox 2 (ENSG00000151615), score: 0.74 PPM1Gprotein phosphatase, Mg2+/Mn2+ dependent, 1G (ENSG00000115241), score: 0.75 PROKR1prokineticin receptor 1 (ENSG00000169618), score: 0.8 PRPHperipherin (ENSG00000135406), score: 0.8 PRSS21protease, serine, 21 (testisin) (ENSG00000007038), score: 0.79 PRSS50protease, serine, 50 (ENSG00000206549), score: 0.78 PRTGprotogenin (ENSG00000166450), score: 0.74 PSAPL1prosaposin-like 1 (gene/pseudogene) (ENSG00000178597), score: 0.74 RAE1RAE1 RNA export 1 homolog (S. pombe) (ENSG00000101146), score: 0.74 RAG2recombination activating gene 2 (ENSG00000175097), score: 0.81 RFX2regulatory factor X, 2 (influences HLA class II expression) (ENSG00000087903), score: 0.74 RNASE11ribonuclease, RNase A family, 11 (non-active) (ENSG00000173464), score: 0.79 RNF113Aring finger protein 113A (ENSG00000125352), score: -0.64 RNF113Bring finger protein 113B (ENSG00000139797), score: 0.84 RPL10Lribosomal protein L10-like (ENSG00000165496), score: 0.76 RSPH6Aradial spoke head 6 homolog A (Chlamydomonas) (ENSG00000104941), score: 0.82 RTP2receptor (chemosensory) transporter protein 2 (ENSG00000198471), score: 0.77 RXFP2relaxin/insulin-like family peptide receptor 2 (ENSG00000133105), score: 0.82 SAGS-antigen; retina and pineal gland (arrestin) (ENSG00000130561), score: 0.81 SMAD7SMAD family member 7 (ENSG00000101665), score: -0.61 SMC1Bstructural maintenance of chromosomes 1B (ENSG00000077935), score: 0.74 SMCPsperm mitochondria-associated cysteine-rich protein (ENSG00000163206), score: 0.74 SPACA1sperm acrosome associated 1 (ENSG00000118434), score: 0.74 SPACA3sperm acrosome associated 3 (ENSG00000141316), score: 0.77 SPACA4sperm acrosome associated 4 (ENSG00000177202), score: 0.76 SPATA19spermatogenesis associated 19 (ENSG00000166118), score: 0.75 SPATA3spermatogenesis associated 3 (ENSG00000173699), score: 0.74 SPDYE4speedy homolog E4 (Xenopus laevis) (ENSG00000183318), score: 0.82 SPERTspermatid associated (ENSG00000174015), score: 0.75 SPO11SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae) (ENSG00000054796), score: 0.82 SPRR4small proline-rich protein 4 (ENSG00000184148), score: 0.86 SPSB4splA/ryanodine receptor domain and SOCS box containing 4 (ENSG00000175093), score: 0.78 STX7syntaxin 7 (ENSG00000079950), score: -0.65 SULT6B1sulfotransferase family, cytosolic, 6B, member 1 (ENSG00000138068), score: 0.87 SYCNsyncollin (ENSG00000179751), score: 0.92 TBX4T-box 4 (ENSG00000121075), score: 0.75 TCP11t-complex 11 homolog (mouse) (ENSG00000124678), score: 0.78 TEX13Atestis expressed 13A (ENSG00000133149), score: 0.79 TEX19testis expressed 19 (ENSG00000182459), score: 0.92 TGM4transglutaminase 4 (prostate) (ENSG00000163810), score: 0.83 TKTL2transketolase-like 2 (ENSG00000151005), score: 0.79 TLX2T-cell leukemia homeobox 2 (ENSG00000115297), score: 0.75 TMCO5Atransmembrane and coiled-coil domains 5A (ENSG00000166069), score: 0.74 TMPRSS12transmembrane (C-terminal) protease, serine 12 (ENSG00000186452), score: 0.76 TP63tumor protein p63 (ENSG00000073282), score: 0.82 TPD52L3tumor protein D52-like 3 (ENSG00000170777), score: 0.83 TREML2triggering receptor expressed on myeloid cells-like 2 (ENSG00000112195), score: 0.86 TRIM31tripartite motif-containing 31 (ENSG00000204616), score: 0.8 TSKStestis-specific serine kinase substrate (ENSG00000126467), score: 0.77 TSPYL6TSPY-like 6 (ENSG00000178021), score: 0.76 TTC19tetratricopeptide repeat domain 19 (ENSG00000011295), score: -0.64 UBQLN3ubiquilin 3 (ENSG00000175520), score: 0.75 VPRBPVpr (HIV-1) binding protein (ENSG00000145041), score: 0.78 VWA3Bvon Willebrand factor A domain containing 3B (ENSG00000168658), score: 0.75 WBSCR28Williams-Beuren syndrome chromosome region 28 (ENSG00000175877), score: 0.76 WDR38WD repeat domain 38 (ENSG00000136918), score: 0.76 WDR69WD repeat domain 69 (ENSG00000123977), score: 0.77 WDR93WD repeat domain 93 (ENSG00000140527), score: 0.8 WFDC11WAP four-disulfide core domain 11 (ENSG00000180083), score: 0.81 WFDC5WAP four-disulfide core domain 5 (ENSG00000175121), score: 0.78 WFDC6WAP four-disulfide core domain 6 (ENSG00000101448), score: 0.74 YSK4YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae) (ENSG00000176601), score: 0.75 ZADH2zinc binding alcohol dehydrogenase domain containing 2 (ENSG00000180011), score: -0.68 ZBTB32zinc finger and BTB domain containing 32 (ENSG00000011590), score: 0.79 ZFP42zinc finger protein 42 homolog (mouse) (ENSG00000179059), score: 0.88 ZMYND10zinc finger, MYND-type containing 10 (ENSG00000004838), score: 0.76 ZNF474zinc finger protein 474 (ENSG00000164185), score: 0.82 ZNF683zinc finger protein 683 (ENSG00000176083), score: 0.74 ZNRF4zinc and ring finger 4 (ENSG00000105428), score: 0.77 ZP1zona pellucida glycoprotein 1 (sperm receptor) (ENSG00000149506), score: 0.78 ZPBP2zona pellucida binding protein 2 (ENSG00000186075), score: 0.76 ZSCAN5Azinc finger and SCAN domain containing 5A (ENSG00000131848), score: 0.8 ZSCAN5Czinc finger and SCAN domain containing 5C (ENSG00000204532), score: 0.83
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_ts_m1_ca1 | hsa | ts | m | 1 |
| hsa_ts_m2_ca1 | hsa | ts | m | 2 |
| ptr_ts_m_ca1 | ptr | ts | m | _ |
| ppa_ts_m_ca1 | ppa | ts | m | _ |