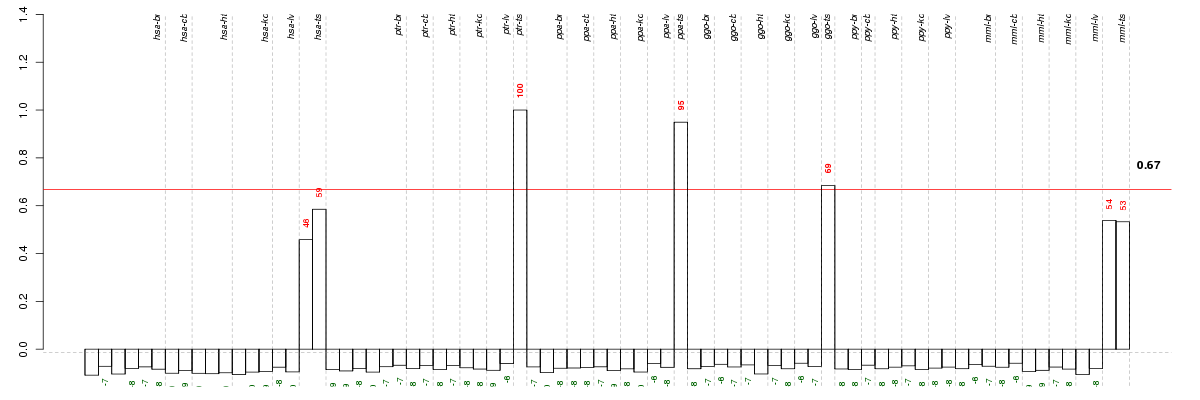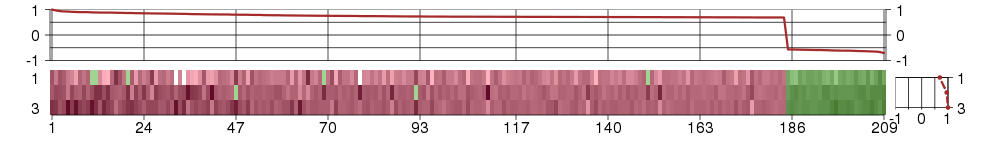

Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
meiosis
A cell cycle process comprising the steps by which a cell progresses through the nuclear division phase of a meiotic cell cycle, the specialized nuclear and cell division in which a single diploid cell undergoes two nuclear divisions following a single round of DNA replication in order to produce four daughter cells that contain half the number of chromosomes as the diploid cell. Meiotic division occurs during the formation of gametes from diploid organisms and at the beginning of haplophase in those organisms that alternate between diploid and haploid generations.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
sexual reproduction
The regular alternation, in the life cycle of haplontic, diplontic and diplohaplontic organisms, of meiosis and fertilization which provides for the production offspring. In diplontic organisms there is a life cycle in which the products of meiosis behave directly as gametes, fusing to form a zygote from which the diploid, or sexually reproductive polyploid, adult organism will develop. In diplohaplontic organisms a haploid phase (gametophyte) exists in the life cycle between meiosis and fertilization (e.g. higher plants, many algae and Fungi); the products of meiosis are spores that develop as haploid individuals from which haploid gametes develop to form a diploid zygote; diplohaplontic organisms show an alternation of haploid and diploid generations. In haplontic organisms meiosis occurs in the zygote, giving rise to four haploid cells (e.g. many algae and protozoa), only the zygote is diploid and this may form a resistant spore, tiding organisms over hard times.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
cell division
The process resulting in the physical partitioning and separation of a cell into daughter cells.
meiotic cell cycle
Progression through the phases of the meiotic cell cycle, in which canonically a cell replicates to produce four offspring with half the chromosomal content of the progenitor cell.
M phase of meiotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the meiotic cell cycle during which meiosis takes place.
all
NA
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
meiosis
A cell cycle process comprising the steps by which a cell progresses through the nuclear division phase of a meiotic cell cycle, the specialized nuclear and cell division in which a single diploid cell undergoes two nuclear divisions following a single round of DNA replication in order to produce four daughter cells that contain half the number of chromosomes as the diploid cell. Meiotic division occurs during the formation of gametes from diploid organisms and at the beginning of haplophase in those organisms that alternate between diploid and haploid generations.
M phase of meiotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the meiotic cell cycle during which meiosis takes place.


AADACL4arylacetamide deacetylase-like 4 (ENSG00000204518), score: 0.76 AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.84 ABCC12ATP-binding cassette, sub-family C (CFTR/MRP), member 12 (ENSG00000140798), score: 0.72 ABLIM3actin binding LIM protein family, member 3 (ENSG00000173210), score: -0.59 ACTL8actin-like 8 (ENSG00000117148), score: 0.75 ADAD1adenosine deaminase domain containing 1 (testis-specific) (ENSG00000164113), score: 0.69 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (ENSG00000129221), score: 0.83 ANKFN1ankyrin-repeat and fibronectin type III domain containing 1 (ENSG00000153930), score: 0.71 APOBEC1apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (ENSG00000111701), score: 0.7 AQP10aquaporin 10 (ENSG00000143595), score: 0.81 ARHGEF10LRho guanine nucleotide exchange factor (GEF) 10-like (ENSG00000074964), score: -0.63 ASPMasp (abnormal spindle) homolog, microcephaly associated (Drosophila) (ENSG00000066279), score: 0.69 AURKCaurora kinase C (ENSG00000105146), score: 0.73 BET3LBET3 like (S. cerevisiae) (ENSG00000173626), score: 0.85 BRCA2breast cancer 2, early onset (ENSG00000139618), score: 0.81 BTG4B-cell translocation gene 4 (ENSG00000137707), score: 0.71 BTN1A1butyrophilin, subfamily 1, member A1 (ENSG00000124557), score: 0.94 C10orf131chromosome 10 open reading frame 131 (ENSG00000173088), score: 0.79 C10orf54chromosome 10 open reading frame 54 (ENSG00000107738), score: -0.61 C11orf88chromosome 11 open reading frame 88 (ENSG00000183644), score: 0.71 C12orf40chromosome 12 open reading frame 40 (ENSG00000180116), score: 0.92 C14orf39chromosome 14 open reading frame 39 (ENSG00000179008), score: 0.79 C17orf50chromosome 17 open reading frame 50 (ENSG00000154768), score: 0.69 C17orf66chromosome 17 open reading frame 66 (ENSG00000172653), score: 0.7 C17orf98chromosome 17 open reading frame 98 (ENSG00000214556), score: 0.71 C1orf14chromosome 1 open reading frame 14 (ENSG00000157060), score: 0.7 C1orf141chromosome 1 open reading frame 141 (ENSG00000203963), score: 0.7 C5orf47chromosome 5 open reading frame 47 (ENSG00000185056), score: 0.72 C5orf50chromosome 5 open reading frame 50 (ENSG00000185662), score: 0.72 C6orf163chromosome 6 open reading frame 163 (ENSG00000203872), score: 0.7 CAPN1calpain 1, (mu/I) large subunit (ENSG00000014216), score: -0.71 CASC5cancer susceptibility candidate 5 (ENSG00000137812), score: 0.78 CATSPER4cation channel, sperm associated 4 (ENSG00000188782), score: 0.71 CC2D2Bcoiled-coil and C2 domain containing 2B (ENSG00000188649), score: 0.77 CCDC105coiled-coil domain containing 105 (ENSG00000160994), score: 0.71 CCDC79coiled-coil domain containing 79 (ENSG00000177461), score: 0.7 CDC20Bcell division cycle 20 homolog B (S. cerevisiae) (ENSG00000164287), score: 0.81 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.72 CENPEcentromere protein E, 312kDa (ENSG00000138778), score: 0.72 CLEC9AC-type lectin domain family 9, member A (ENSG00000197992), score: 0.75 CLSPNclaspin (ENSG00000092853), score: 0.88 COMMD7COMM domain containing 7 (ENSG00000149600), score: -0.67 COPS7ACOP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) (ENSG00000111652), score: -0.59 CSN3casein kappa (ENSG00000171209), score: 0.7 CSTL1cystatin-like 1 (ENSG00000125823), score: 0.76 CYLC2cylicin, basic protein of sperm head cytoskeleton 2 (ENSG00000155833), score: 0.71 DBF4DBF4 homolog (S. cerevisiae) (ENSG00000006634), score: 0.7 DKFZP781G0119hypothetical protein LOC644041 (ENSG00000206043), score: 0.75 DKK4dickkopf homolog 4 (Xenopus laevis) (ENSG00000104371), score: 0.86 DMP1dentin matrix acidic phosphoprotein 1 (ENSG00000152592), score: 0.83 DMRT3doublesex and mab-3 related transcription factor 3 (ENSG00000064218), score: 0.73 DMRTB1DMRT-like family B with proline-rich C-terminal, 1 (ENSG00000143006), score: 0.72 DRG1developmentally regulated GTP binding protein 1 (ENSG00000185721), score: 0.71 DTLdenticleless homolog (Drosophila) (ENSG00000143476), score: 0.69 E2F2E2F transcription factor 2 (ENSG00000007968), score: 0.73 E2F7E2F transcription factor 7 (ENSG00000165891), score: 0.74 ELL3elongation factor RNA polymerase II-like 3 (ENSG00000128886), score: 0.69 ENTHD1ENTH domain containing 1 (ENSG00000176177), score: 0.69 EVX1even-skipped homeobox 1 (ENSG00000106038), score: 0.73 EXD1exonuclease 3'-5' domain containing 1 (ENSG00000178997), score: 0.71 FAM177Bfamily with sequence similarity 177, member B (ENSG00000197520), score: 0.88 FAM47Bfamily with sequence similarity 47, member B (ENSG00000189132), score: 0.72 FAM47Cfamily with sequence similarity 47, member C (ENSG00000198173), score: 0.76 FAM55Afamily with sequence similarity 55, member A (ENSG00000095110), score: 0.74 FAM65Afamily with sequence similarity 65, member A (ENSG00000039523), score: -0.58 FAM9Cfamily with sequence similarity 9, member C (ENSG00000187268), score: 0.9 FBXO47F-box protein 47 (ENSG00000204952), score: 0.7 FER1L6fer-1-like 6 (C. elegans) (ENSG00000214814), score: 0.7 FGF4fibroblast growth factor 4 (ENSG00000075388), score: 0.82 FLOT2flotillin 2 (ENSG00000132589), score: -0.61 FOXN4forkhead box N4 (ENSG00000139445), score: 0.69 FSIP2fibrous sheath interacting protein 2 (ENSG00000188738), score: 0.7 G6PC2glucose-6-phosphatase, catalytic, 2 (ENSG00000152254), score: 0.79 GAB4GRB2-associated binding protein family, member 4 (ENSG00000215568), score: 0.82 GATA1GATA binding protein 1 (globin transcription factor 1) (ENSG00000102145), score: 0.85 GBX1gastrulation brain homeobox 1 (ENSG00000164900), score: 0.77 GJA8gap junction protein, alpha 8, 50kDa (ENSG00000121634), score: 0.84 GPR156G protein-coupled receptor 156 (ENSG00000175697), score: 0.78 GRXCR2glutaredoxin, cysteine rich 2 (ENSG00000204928), score: 0.84 GUCY2Dguanylate cyclase 2D, membrane (retina-specific) (ENSG00000132518), score: 0.75 HDAC8histone deacetylase 8 (ENSG00000147099), score: -0.57 HMMRhyaluronan-mediated motility receptor (RHAMM) (ENSG00000072571), score: 0.72 HORMAD1HORMA domain containing 1 (ENSG00000143452), score: 0.69 HOXB13homeobox B13 (ENSG00000159184), score: 0.75 IDH3Gisocitrate dehydrogenase 3 (NAD+) gamma (ENSG00000067829), score: -0.61 IGF2BP3insulin-like growth factor 2 mRNA binding protein 3 (ENSG00000136231), score: 0.71 IL13interleukin 13 (ENSG00000169194), score: 0.76 IL1F5interleukin 1 family, member 5 (delta) (ENSG00000136695), score: 0.81 IL23Rinterleukin 23 receptor (ENSG00000162594), score: 0.69 IL31RAinterleukin 31 receptor A (ENSG00000164509), score: 0.77 INSL6insulin-like 6 (ENSG00000120210), score: 0.72 KIF20Bkinesin family member 20B (ENSG00000138182), score: 0.72 KLF17Kruppel-like factor 17 (ENSG00000171872), score: 0.71 KLF9Kruppel-like factor 9 (ENSG00000119138), score: -0.65 KLK12kallikrein-related peptidase 12 (ENSG00000186474), score: 0.88 KLK13kallikrein-related peptidase 13 (ENSG00000167759), score: 0.83 KLRC4killer cell lectin-like receptor subfamily C, member 4 (ENSG00000183542), score: 0.72 KRT1keratin 1 (ENSG00000167768), score: 0.86 KRT26keratin 26 (ENSG00000186393), score: 0.77 KRT79keratin 79 (ENSG00000185640), score: 0.74 L1TD1LINE-1 type transposase domain containing 1 (ENSG00000240563), score: 0.72 LCTLlactase-like (ENSG00000188501), score: 0.78 LEKR1leucine, glutamate and lysine rich 1 (ENSG00000197980), score: 0.69 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.82 LIN28Alin-28 homolog A (C. elegans) (ENSG00000131914), score: 0.75 LIPT1lipoyltransferase 1 (ENSG00000144182), score: 0.7 LOC100292575similar to melanoma antigen family C, 2 (ENSG00000046774), score: 0.7 LOXHD1lipoxygenase homology domains 1 (ENSG00000167210), score: 0.75 LRIT1leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 (ENSG00000148602), score: 0.84 LRRC8Aleucine rich repeat containing 8 family, member A (ENSG00000136802), score: -0.57 LRRCC1leucine rich repeat and coiled-coil domain containing 1 (ENSG00000133739), score: 0.7 LRRIQ1leucine-rich repeats and IQ motif containing 1 (ENSG00000133640), score: 0.7 MAGEA4melanoma antigen family A, 4 (ENSG00000147381), score: 0.7 MAGEB10melanoma antigen family B, 10 (ENSG00000177689), score: 0.69 MAGED2melanoma antigen family D, 2 (ENSG00000102316), score: -0.61 MARCH9membrane-associated ring finger (C3HC4) 9 (ENSG00000139266), score: -0.59 MEP1Ameprin A, alpha (PABA peptide hydrolase) (ENSG00000112818), score: 0.88 MGLLmonoglyceride lipase (ENSG00000074416), score: -0.64 MLF2myeloid leukemia factor 2 (ENSG00000089693), score: -0.57 MS4A12membrane-spanning 4-domains, subfamily A, member 12 (ENSG00000071203), score: 1 MS4A6Emembrane-spanning 4-domains, subfamily A, member 6E (ENSG00000166926), score: 0.76 MYO3Amyosin IIIA (ENSG00000095777), score: 0.69 NAA10N(alpha)-acetyltransferase 10, NatA catalytic subunit (ENSG00000102030), score: -0.62 NENFneuron derived neurotrophic factor (ENSG00000117691), score: -0.58 NEUROD4neurogenic differentiation 4 (ENSG00000123307), score: 0.8 NMBRneuromedin B receptor (ENSG00000135577), score: 0.81 NUP210Lnucleoporin 210kDa-like (ENSG00000143552), score: 0.7 ODF4outer dense fiber of sperm tails 4 (ENSG00000184650), score: 0.7 OR2B3olfactory receptor, family 2, subfamily B, member 3 (ENSG00000204703), score: 0.8 OR51T1olfactory receptor, family 51, subfamily T, member 1 (ENSG00000176900), score: 0.73 OR56A1olfactory receptor, family 56, subfamily A, member 1 (ENSG00000180934), score: 0.71 ORC1Lorigin recognition complex, subunit 1-like (S. cerevisiae) (ENSG00000085840), score: 0.71 PADI1peptidyl arginine deiminase, type I (ENSG00000142623), score: 0.88 PARP11poly (ADP-ribose) polymerase family, member 11 (ENSG00000111224), score: 0.71 PBKPDZ binding kinase (ENSG00000168078), score: 0.73 PDE6Aphosphodiesterase 6A, cGMP-specific, rod, alpha (ENSG00000132915), score: 0.91 PDE6Hphosphodiesterase 6H, cGMP-specific, cone, gamma (ENSG00000139053), score: 0.8 PDHA2pyruvate dehydrogenase (lipoamide) alpha 2 (ENSG00000163114), score: 0.71 PDZD11PDZ domain containing 11 (ENSG00000120509), score: -0.57 PGLYRP3peptidoglycan recognition protein 3 (ENSG00000159527), score: 0.92 PGLYRP4peptidoglycan recognition protein 4 (ENSG00000163218), score: 0.84 PLA2G2Ephospholipase A2, group IIE (ENSG00000188784), score: 0.89 PLCZ1phospholipase C, zeta 1 (ENSG00000139151), score: 0.72 PLSCR2phospholipid scramblase 2 (ENSG00000163746), score: 0.7 POM121L2POM121 membrane glycoprotein-like 2 (ENSG00000158553), score: 0.71 PPFIA1protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 (ENSG00000131626), score: 0.7 PRTGprotogenin (ENSG00000166450), score: 0.77 RAE1RAE1 RNA export 1 homolog (S. pombe) (ENSG00000101146), score: 0.71 RAG2recombination activating gene 2 (ENSG00000175097), score: 0.86 RBM46RNA binding motif protein 46 (ENSG00000151962), score: 0.71 RNASE11ribonuclease, RNase A family, 11 (non-active) (ENSG00000173464), score: 0.72 RNF113Aring finger protein 113A (ENSG00000125352), score: -0.6 RNF113Bring finger protein 113B (ENSG00000139797), score: 0.75 RPP38ribonuclease P/MRP 38kDa subunit (ENSG00000152464), score: 0.7 RSPH4Aradial spoke head 4 homolog A (Chlamydomonas) (ENSG00000111834), score: 0.69 RSPH6Aradial spoke head 6 homolog A (Chlamydomonas) (ENSG00000104941), score: 0.7 RTKN2rhotekin 2 (ENSG00000182010), score: 0.75 RXFP2relaxin/insulin-like family peptide receptor 2 (ENSG00000133105), score: 0.9 SAGS-antigen; retina and pineal gland (arrestin) (ENSG00000130561), score: 0.8 SCAND3SCAN domain containing 3 (ENSG00000232040), score: 0.69 SENP2SUMO1/sentrin/SMT3 specific peptidase 2 (ENSG00000163904), score: 0.74 SEPT14septin 14 (ENSG00000154997), score: 0.71 SGOL2shugoshin-like 2 (S. pombe) (ENSG00000163535), score: 0.69 SIsucrase-isomaltase (alpha-glucosidase) (ENSG00000090402), score: 0.76 SLC25A31solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 (ENSG00000151475), score: 0.71 SMAD7SMAD family member 7 (ENSG00000101665), score: -0.65 SMC1Bstructural maintenance of chromosomes 1B (ENSG00000077935), score: 0.71 SPDYE4speedy homolog E4 (Xenopus laevis) (ENSG00000183318), score: 0.73 SPO11SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae) (ENSG00000054796), score: 0.79 SPRR4small proline-rich protein 4 (ENSG00000184148), score: 0.84 SPSB4splA/ryanodine receptor domain and SOCS box containing 4 (ENSG00000175093), score: 0.7 SULT6B1sulfotransferase family, cytosolic, 6B, member 1 (ENSG00000138068), score: 0.83 SYCP1synaptonemal complex protein 1 (ENSG00000198765), score: 0.73 TBC1D17TBC1 domain family, member 17 (ENSG00000104946), score: -0.61 TBX22T-box 22 (ENSG00000122145), score: 0.71 TEX15testis expressed 15 (ENSG00000133863), score: 0.71 TEX19testis expressed 19 (ENSG00000182459), score: 0.8 TGM4transglutaminase 4 (prostate) (ENSG00000163810), score: 0.96 TKTL2transketolase-like 2 (ENSG00000151005), score: 0.72 TREML2triggering receptor expressed on myeloid cells-like 2 (ENSG00000112195), score: 0.72 TRIM31tripartite motif-containing 31 (ENSG00000204616), score: 0.9 UBL4Aubiquitin-like 4A (ENSG00000102178), score: -0.59 USP44ubiquitin specific peptidase 44 (ENSG00000136014), score: 0.69 VASH2vasohibin 2 (ENSG00000143494), score: 0.69 VPRBPVpr (HIV-1) binding protein (ENSG00000145041), score: 0.71 WDR45WD repeat domain 45 (ENSG00000196998), score: -0.63 WDR69WD repeat domain 69 (ENSG00000123977), score: 0.7 WDR93WD repeat domain 93 (ENSG00000140527), score: 0.74 WFDC11WAP four-disulfide core domain 11 (ENSG00000180083), score: 0.9 WFDC5WAP four-disulfide core domain 5 (ENSG00000175121), score: 0.72 WISP3WNT1 inducible signaling pathway protein 3 (ENSG00000112761), score: 0.71 ZADH2zinc binding alcohol dehydrogenase domain containing 2 (ENSG00000180011), score: -0.63 ZFP42zinc finger protein 42 homolog (mouse) (ENSG00000179059), score: 0.73 ZNF474zinc finger protein 474 (ENSG00000164185), score: 0.69 ZNF572zinc finger protein 572 (ENSG00000180938), score: 0.7 ZP1zona pellucida glycoprotein 1 (sperm receptor) (ENSG00000149506), score: 0.74 ZPBP2zona pellucida binding protein 2 (ENSG00000186075), score: 0.69 ZSCAN5Czinc finger and SCAN domain containing 5C (ENSG00000204532), score: 0.72
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_ts_m_ca1 | ggo | ts | m | _ |
| ppa_ts_m_ca1 | ppa | ts | m | _ |
| ptr_ts_m_ca1 | ptr | ts | m | _ |