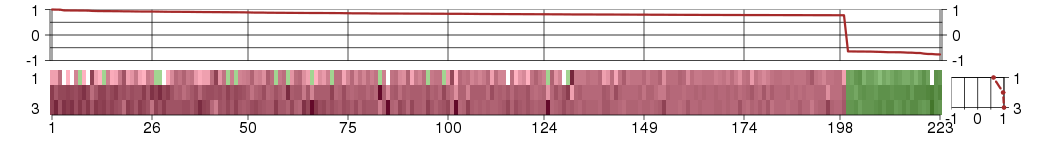

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
sensory perception
The series of events required for an organism to receive a sensory stimulus, convert it to a molecular signal, and recognize and characterize the signal. This is a neurological process.
sensory perception of chemical stimulus
The series of events required for an organism to receive a sensory chemical stimulus, convert it to a molecular signal, and recognize and characterize the signal. This is a neurological process.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
all
NA


ACSM4acyl-CoA synthetase medium-chain family member 4 (ENSG00000215009), score: 0.94 ADAM32ADAM metallopeptidase domain 32 (ENSG00000197140), score: 0.79 ADAMTS20ADAM metallopeptidase with thrombospondin type 1 motif, 20 (ENSG00000173157), score: 0.84 AGBL3ATP/GTP binding protein-like 3 (ENSG00000146856), score: 0.84 AK7adenylate kinase 7 (ENSG00000140057), score: 0.81 AKD1adenylate kinase domain containing 1 (ENSG00000155085), score: 0.79 AKNAD1AKNA domain containing 1 (ENSG00000162641), score: 0.84 ANKRD60ankyrin repeat domain 60 (ENSG00000124227), score: 0.89 ANKRD62ankyrin repeat domain 62 (ENSG00000181626), score: 0.8 APLP2amyloid beta (A4) precursor-like protein 2 (ENSG00000084234), score: -0.64 APOBEC4apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) (ENSG00000173627), score: 0.83 ARL13AADP-ribosylation factor-like 13A (ENSG00000174225), score: 0.84 ASB5ankyrin repeat and SOCS box-containing 5 (ENSG00000164122), score: 0.9 BEND2BEN domain containing 2 (ENSG00000177324), score: 0.78 C10orf120chromosome 10 open reading frame 120 (ENSG00000183559), score: 0.86 C10orf122chromosome 10 open reading frame 122 (ENSG00000175018), score: 0.8 C10orf67chromosome 10 open reading frame 67 (ENSG00000179133), score: 0.85 C11orf16chromosome 11 open reading frame 16 (ENSG00000176029), score: 0.92 C11orf82chromosome 11 open reading frame 82 (ENSG00000165490), score: 0.79 C11orf83chromosome 11 open reading frame 83 (ENSG00000204922), score: -0.65 C12orf56chromosome 12 open reading frame 56 (ENSG00000185306), score: 0.78 C12orf63chromosome 12 open reading frame 63 (ENSG00000188596), score: 0.82 C12orf71chromosome 12 open reading frame 71 (ENSG00000214700), score: 0.84 C14orf145chromosome 14 open reading frame 145 (ENSG00000100629), score: 0.8 C18orf21chromosome 18 open reading frame 21 (ENSG00000141428), score: 0.78 C18orf34chromosome 18 open reading frame 34 (ENSG00000166960), score: 0.81 C1orf129chromosome 1 open reading frame 129 (ENSG00000117501), score: 0.93 C1orf141chromosome 1 open reading frame 141 (ENSG00000203963), score: 0.81 C1orf185chromosome 1 open reading frame 185 (ENSG00000204006), score: 0.8 C1orf87chromosome 1 open reading frame 87 (ENSG00000162598), score: 0.82 C20orf114chromosome 20 open reading frame 114 (ENSG00000125999), score: 0.92 C20orf173chromosome 20 open reading frame 173 (ENSG00000125975), score: 0.79 C20orf185chromosome 20 open reading frame 185 (ENSG00000186190), score: 0.83 C22orf31chromosome 22 open reading frame 31 (ENSG00000100249), score: 0.83 C3orf22chromosome 3 open reading frame 22 (ENSG00000180697), score: 0.8 C3orf77chromosome 3 open reading frame 77 (ENSG00000173769), score: 0.8 C4orf17chromosome 4 open reading frame 17 (ENSG00000138813), score: 0.78 C5orf48chromosome 5 open reading frame 48 (ENSG00000196900), score: 0.84 C6orf103chromosome 6 open reading frame 103 (ENSG00000118492), score: 0.78 C6orf25chromosome 6 open reading frame 25 (ENSG00000204420), score: 0.78 C7orf57chromosome 7 open reading frame 57 (ENSG00000164746), score: 0.83 C9orf135chromosome 9 open reading frame 135 (ENSG00000204711), score: 0.81 C9orf50chromosome 9 open reading frame 50 (ENSG00000179058), score: 0.9 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.86 CCDC15coiled-coil domain containing 15 (ENSG00000149548), score: 0.82 CCDC22coiled-coil domain containing 22 (ENSG00000101997), score: -0.71 CCDC38coiled-coil domain containing 38 (ENSG00000165972), score: 0.83 CCDC60coiled-coil domain containing 60 (ENSG00000183273), score: 0.78 CCDC63coiled-coil domain containing 63 (ENSG00000173093), score: 0.78 CCDC67coiled-coil domain containing 67 (ENSG00000165325), score: 0.78 CCDC83coiled-coil domain containing 83 (ENSG00000150676), score: 0.79 CD200R1LCD200 receptor 1-like (ENSG00000206531), score: 0.88 CD96CD96 molecule (ENSG00000153283), score: 0.83 CDCA7cell division cycle associated 7 (ENSG00000144354), score: 0.79 CDKL3cyclin-dependent kinase-like 3 (ENSG00000006837), score: 0.8 CDKL4cyclin-dependent kinase-like 4 (ENSG00000205111), score: 0.84 CENPFcentromere protein F, 350/400kDa (mitosin) (ENSG00000117724), score: 0.8 CEP152centrosomal protein 152kDa (ENSG00000103995), score: 0.78 CHIAchitinase, acidic (ENSG00000134216), score: 0.84 CKAP2Lcytoskeleton associated protein 2-like (ENSG00000169607), score: 0.79 CLCA2chloride channel accessory 2 (ENSG00000137975), score: 0.87 CNBD1cyclic nucleotide binding domain containing 1 (ENSG00000176571), score: 0.91 CNGA3cyclic nucleotide gated channel alpha 3 (ENSG00000144191), score: 0.89 CNOT10CCR4-NOT transcription complex, subunit 10 (ENSG00000182973), score: 0.82 CRYBA4crystallin, beta A4 (ENSG00000196431), score: 0.77 CST8cystatin 8 (cystatin-related epididymal specific) (ENSG00000125815), score: 0.8 CST9cystatin 9 (testatin) (ENSG00000173335), score: 0.96 CSTL1cystatin-like 1 (ENSG00000125823), score: 0.8 CTBP1C-terminal binding protein 1 (ENSG00000159692), score: -0.68 CTLA4cytotoxic T-lymphocyte-associated protein 4 (ENSG00000163599), score: 0.91 CXorf22chromosome X open reading frame 22 (ENSG00000165164), score: 0.99 CXorf30chromosome X open reading frame 30 (ENSG00000205081), score: 0.91 CXorf41chromosome X open reading frame 41 (ENSG00000080572), score: 0.8 CYLC1cylicin, basic protein of sperm head cytoskeleton 1 (ENSG00000183035), score: 0.78 CYP19A1cytochrome P450, family 19, subfamily A, polypeptide 1 (ENSG00000137869), score: 0.82 DCDC1doublecortin domain containing 1 (ENSG00000188682), score: 0.85 DEFB135defensin, beta 135 (ENSG00000205883), score: 0.93 DLGAP5discs, large (Drosophila) homolog-associated protein 5 (ENSG00000126787), score: 0.79 DNAH6dynein, axonemal, heavy chain 6 (ENSG00000115423), score: 0.79 DNAJB7DnaJ (Hsp40) homolog, subfamily B, member 7 (ENSG00000172404), score: 0.78 E2F8E2F transcription factor 8 (ENSG00000129173), score: 0.82 EFCAB5EF-hand calcium binding domain 5 (ENSG00000176927), score: 0.78 EFCAB9EF-hand calcium binding domain 9 (ENSG00000214360), score: 0.8 EFHBEF-hand domain family, member B (ENSG00000163576), score: 0.79 ERCC6Lexcision repair cross-complementing rodent repair deficiency, complementation group 6-like (ENSG00000186871), score: 0.88 ESCO2establishment of cohesion 1 homolog 2 (S. cerevisiae) (ENSG00000171320), score: 0.79 FAM194Afamily with sequence similarity 194, member A (ENSG00000163645), score: 0.78 FAM46Dfamily with sequence similarity 46, member D (ENSG00000174016), score: 0.87 FAM50Afamily with sequence similarity 50, member A (ENSG00000071859), score: -0.68 FCRL4Fc receptor-like 4 (ENSG00000163518), score: 0.91 FKBPLFK506 binding protein like (ENSG00000204315), score: 0.78 FLOT1flotillin 1 (ENSG00000137312), score: -0.75 FOXB1forkhead box B1 (ENSG00000171956), score: 0.8 GDPD4glycerophosphodiester phosphodiesterase domain containing 4 (ENSG00000178795), score: 0.8 GLIPR1L1GLI pathogenesis-related 1 like 1 (ENSG00000173401), score: 0.89 GLT6D1glycosyltransferase 6 domain containing 1 (ENSG00000204007), score: 0.92 GPA33glycoprotein A33 (transmembrane) (ENSG00000143167), score: 0.81 GPR144G protein-coupled receptor 144 (ENSG00000180264), score: 0.92 GPR18G protein-coupled receptor 18 (ENSG00000125245), score: 0.79 GRXCR1glutaredoxin, cysteine rich 1 (ENSG00000215203), score: 0.9 GSG2germ cell associated 2 (haspin) (ENSG00000177602), score: 0.81 GUCY2Cguanylate cyclase 2C (heat stable enterotoxin receptor) (ENSG00000070019), score: 0.78 GUCY2Fguanylate cyclase 2F, retinal (ENSG00000101890), score: 0.94 H1FOOH1 histone family, member O, oocyte-specific (ENSG00000178804), score: 0.78 H2AFYH2A histone family, member Y (ENSG00000113648), score: -0.68 HDChistidine decarboxylase (ENSG00000140287), score: 0.85 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (ENSG00000126945), score: -0.64 HOXA13homeobox A13 (ENSG00000106031), score: 0.93 IFLTD1intermediate filament tail domain containing 1 (ENSG00000152936), score: 0.91 IGF2BP3insulin-like growth factor 2 mRNA binding protein 3 (ENSG00000136231), score: 0.78 IL19interleukin 19 (ENSG00000142224), score: 0.86 IL1Ainterleukin 1, alpha (ENSG00000115008), score: 0.83 IL20interleukin 20 (ENSG00000162891), score: 0.82 IL31interleukin 31 (ENSG00000204671), score: 0.92 IL4interleukin 4 (ENSG00000113520), score: 0.79 IQCHIQ motif containing H (ENSG00000103599), score: 0.78 JDP2Jun dimerization protein 2 (ENSG00000140044), score: -0.69 KCNK18potassium channel, subfamily K, member 18 (ENSG00000186795), score: 0.9 KCNU1potassium channel, subfamily U, member 1 (ENSG00000215262), score: 0.8 KIAA0586KIAA0586 (ENSG00000100578), score: 0.81 KIF14kinesin family member 14 (ENSG00000118193), score: 0.79 KIF18Akinesin family member 18A (ENSG00000121621), score: 0.78 KRT75keratin 75 (ENSG00000170454), score: 0.95 LCTlactase (ENSG00000115850), score: 0.88 LEKR1leucine, glutamate and lysine rich 1 (ENSG00000197980), score: 0.81 LHCGRluteinizing hormone/choriogonadotropin receptor (ENSG00000138039), score: 0.82 LRRC63leucine rich repeat containing 63 (ENSG00000173988), score: 0.78 LRRC67leucine rich repeat containing 67 (ENSG00000178125), score: 0.87 LRRC69leucine rich repeat containing 69 (ENSG00000214954), score: 0.86 LRRIQ4leucine-rich repeats and IQ motif containing 4 (ENSG00000188306), score: 0.89 MAGEA8melanoma antigen family A, 8 (ENSG00000156009), score: 0.94 MC2Rmelanocortin 2 receptor (adrenocorticotropic hormone) (ENSG00000185231), score: 0.9 MELKmaternal embryonic leucine zipper kinase (ENSG00000165304), score: 0.82 MEP1Bmeprin A, beta (ENSG00000141434), score: 0.89 MOSv-mos Moloney murine sarcoma viral oncogene homolog (ENSG00000172680), score: 0.91 MPP1membrane protein, palmitoylated 1, 55kDa (ENSG00000130830), score: -0.69 MTNR1Bmelatonin receptor 1B (ENSG00000134640), score: 0.93 MUC16mucin 16, cell surface associated (ENSG00000181143), score: 0.96 MUC21mucin 21, cell surface associated (ENSG00000204544), score: 0.83 MUC7mucin 7, secreted (ENSG00000171195), score: 0.84 MYH13myosin, heavy chain 13, skeletal muscle (ENSG00000006788), score: 0.94 MYH15myosin, heavy chain 15 (ENSG00000144821), score: 0.86 MYO1Amyosin IA (ENSG00000166866), score: 0.87 NAA10N(alpha)-acetyltransferase 10, NatA catalytic subunit (ENSG00000102030), score: -0.65 NCAPGnon-SMC condensin I complex, subunit G (ENSG00000109805), score: 0.78 NLRP13NLR family, pyrin domain containing 13 (ENSG00000173572), score: 0.78 NT5DC45'-nucleotidase domain containing 4 (ENSG00000144130), score: 0.86 OR10G3olfactory receptor, family 10, subfamily G, member 3 (ENSG00000169208), score: 0.97 OR13G1olfactory receptor, family 13, subfamily G, member 1 (ENSG00000197437), score: 1 OR4K1olfactory receptor, family 4, subfamily K, member 1 (ENSG00000155249), score: 0.83 OR51V1olfactory receptor, family 51, subfamily V, member 1 (ENSG00000176742), score: 0.86 OR9K2olfactory receptor, family 9, subfamily K, member 2 (ENSG00000170605), score: 1 PAGE3P antigen family, member 3 (prostate associated) (ENSG00000204279), score: 0.83 PCDHB1protocadherin beta 1 (ENSG00000171815), score: 0.96 PGK1phosphoglycerate kinase 1 (ENSG00000102144), score: -0.65 PLA2G2Cphospholipase A2, group IIC (ENSG00000187980), score: 0.9 PLAC1Lplacenta-specific 1-like (ENSG00000149507), score: 0.85 PLK1S1polo-like kinase 1 substrate 1 (ENSG00000088970), score: 0.82 POLQpolymerase (DNA directed), theta (ENSG00000051341), score: 0.84 PQBP1polyglutamine binding protein 1 (ENSG00000102103), score: -0.76 PRDX2peroxiredoxin 2 (ENSG00000167815), score: -0.74 PRR23Bproline rich 23B (ENSG00000184814), score: 0.9 PRSS48protease, serine, 48 (ENSG00000189099), score: 0.82 PTCHD3patched domain containing 3 (ENSG00000182077), score: 0.83 RAB24RAB24, member RAS oncogene family (ENSG00000169228), score: -0.66 RAD51AP1RAD51 associated protein 1 (ENSG00000111247), score: 0.8 RALBP1ralA binding protein 1 (ENSG00000017797), score: -0.66 RBM10RNA binding motif protein 10 (ENSG00000182872), score: -0.68 RP1retinitis pigmentosa 1 (autosomal dominant) (ENSG00000104237), score: 0.96 RPE65retinal pigment epithelium-specific protein 65kDa (ENSG00000116745), score: 0.82 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (ENSG00000092200), score: 0.78 SCELsciellin (ENSG00000136155), score: 0.96 SERPINB12serpin peptidase inhibitor, clade B (ovalbumin), member 12 (ENSG00000166634), score: 0.91 SERPINB5serpin peptidase inhibitor, clade B (ovalbumin), member 5 (ENSG00000206075), score: 0.78 SIX1SIX homeobox 1 (ENSG00000126778), score: 0.81 SLC5A4solute carrier family 5 (low affinity glucose cotransporter), member 4 (ENSG00000100191), score: 0.81 SLC9A10solute carrier family 9, member 10 (ENSG00000172139), score: 0.87 SLC9A11solute carrier family 9, member 11 (ENSG00000162753), score: 0.92 SLCO6A1solute carrier organic anion transporter family, member 6A1 (ENSG00000205359), score: 0.78 SNX12sorting nexin 12 (ENSG00000147164), score: -0.71 SNX21sorting nexin family member 21 (ENSG00000124104), score: -0.65 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (ENSG00000106304), score: 0.86 SPC25SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000152253), score: 0.8 TAS1R2taste receptor, type 1, member 2 (ENSG00000179002), score: 0.96 TAS2R16taste receptor, type 2, member 16 (ENSG00000128519), score: 0.81 TFAP2Dtranscription factor AP-2 delta (activating enhancer binding protein 2 delta) (ENSG00000008197), score: 0.93 TGM7transglutaminase 7 (ENSG00000159495), score: 0.89 TIGD4tigger transposable element derived 4 (ENSG00000169989), score: 0.92 TMC5transmembrane channel-like 5 (ENSG00000103534), score: 0.84 TMEM225transmembrane protein 225 (ENSG00000204300), score: 0.83 TMPRSS7transmembrane protease, serine 7 (ENSG00000176040), score: 0.87 TNP2transition protein 2 (during histone to protamine replacement) (ENSG00000178279), score: 0.84 TRIML1tripartite motif family-like 1 (ENSG00000184108), score: 0.79 TRIML2tripartite motif family-like 2 (ENSG00000179046), score: 0.79 TRYX3trypsin X3 (ENSG00000171147), score: 0.8 TSHBthyroid stimulating hormone, beta (ENSG00000134200), score: 0.86 TTLL8tubulin tyrosine ligase-like family, member 8 (ENSG00000138892), score: 0.81 TXNDC3thioredoxin domain containing 3 (spermatozoa) (ENSG00000086288), score: 0.8 TXNDC8thioredoxin domain containing 8 (spermatozoa) (ENSG00000204193), score: 0.86 TYK2tyrosine kinase 2 (ENSG00000105397), score: -0.65 UBE2Uubiquitin-conjugating enzyme E2U (putative) (ENSG00000177414), score: 0.78 UCP1uncoupling protein 1 (mitochondrial, proton carrier) (ENSG00000109424), score: 0.93 UPRTuracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) (ENSG00000094841), score: -0.68 USF1upstream transcription factor 1 (ENSG00000158773), score: -0.67 USP26ubiquitin specific peptidase 26 (ENSG00000134588), score: 0.78 USP50ubiquitin specific peptidase 50 (ENSG00000170236), score: 0.88 VPREB1pre-B lymphocyte 1 (ENSG00000169575), score: 0.87 WDR45WD repeat domain 45 (ENSG00000196998), score: -0.73 WDR49WD repeat domain 49 (ENSG00000174776), score: 0.89 WDR64WD repeat domain 64 (ENSG00000162843), score: 0.84 XAGE5X antigen family, member 5 (ENSG00000171405), score: 0.78 XKR5XK, Kell blood group complex subunit-related family, member 5 (ENSG00000186530), score: 0.84 ZAR1zygote arrest 1 (ENSG00000182223), score: 0.86 ZNF358zinc finger protein 358 (ENSG00000198816), score: -0.76 ZNF416zinc finger protein 416 (ENSG00000083817), score: 0.85 ZNF560zinc finger protein 560 (ENSG00000198028), score: 0.84 ZNF677zinc finger protein 677 (ENSG00000197928), score: 0.79 ZPLD1zona pellucida-like domain containing 1 (ENSG00000170044), score: 0.87 ZSWIM2zinc finger, SWIM-type containing 2 (ENSG00000163012), score: 0.82
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_ts_m_ca1 | ptr | ts | m | _ |
| mml_ts_m2_ca1 | mml | ts | m | 2 |
| mml_ts_m1_ca1 | mml | ts | m | 1 |