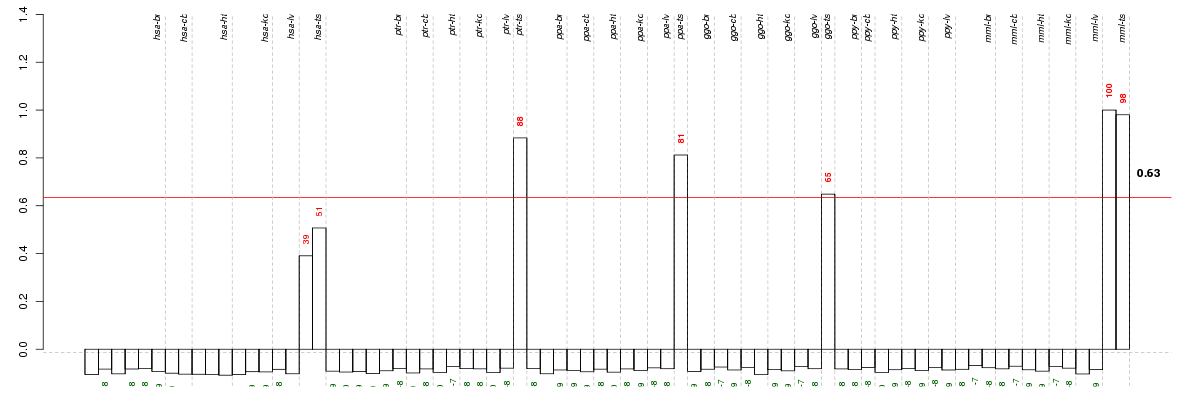

Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
chromosome segregation
The process by which genetic material, in the form of chromosomes, is organized into specific structures and then physically separated and apportioned to two or more sets.
chromosome organization
A process that is carried out at the cellular level that results in the assembly, arrangement of constituent parts, or disassembly of chromosomes, structures composed of a very long molecule of DNA and associated proteins that carries hereditary information.
mitotic sister chromatid segregation
The cell cycle process whereby replicated homologous chromosomes are organized and then physically separated and apportioned to two sets during the mitotic cell cycle. Each replicated chromosome, composed of two sister chromatids, aligns at the cell equator, paired with its homologous partner. One homolog of each morphologic type goes into each of the resulting chromosome sets.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
nuclear division
A process by which a cell nucleus is divided into two nuclei, with DNA and other nuclear contents distributed between the daughter nuclei.
sister chromatid segregation
The process by which sister chromatids are organized and then physically separated and apportioned to two or more sets.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
cellular component movement
The directed, self-propelled movement of a cellular component without the involvement of an external agent such as a transporter or a pore.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
mitotic chromosome movement towards spindle pole
The cell cycle process whereby the directed movement of chromosomes from the center of the spindle towards the spindle poles occurs. This mediates by the shortening of microtubules attached to the chromosomes, during mitosis.
regulation of mitosis
Any process that modulates the frequency, rate or extent of mitosis.
mitotic metaphase/anaphase transition
The cell cycle process whereby a cell progresses from metaphase to anaphase during mitosis, triggered by the destruction of mitotic cyclins.
meiosis
A cell cycle process comprising the steps by which a cell progresses through the nuclear division phase of a meiotic cell cycle, the specialized nuclear and cell division in which a single diploid cell undergoes two nuclear divisions following a single round of DNA replication in order to produce four daughter cells that contain half the number of chromosomes as the diploid cell. Meiotic division occurs during the formation of gametes from diploid organisms and at the beginning of haplophase in those organisms that alternate between diploid and haploid generations.
gamete generation
The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell.
spermatogenesis
The process of formation of spermatozoa, including spermatocytogenesis and spermiogenesis.
regulation of mitotic cell cycle
Any process that modulates the rate or extent of progress through the mitotic cell cycle.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
fertilization
The union of gametes of opposite sexes during the process of sexual reproduction to form a zygote. It involves the fusion of the gametic nuclei (karyogamy) and cytoplasm (plasmogamy).
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cell cycle process
Any process that modulates a cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
positive regulation of organelle organization
Any process that increases the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of an organelle.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
sexual reproduction
The regular alternation, in the life cycle of haplontic, diplontic and diplohaplontic organisms, of meiosis and fertilization which provides for the production offspring. In diplontic organisms there is a life cycle in which the products of meiosis behave directly as gametes, fusing to form a zygote from which the diploid, or sexually reproductive polyploid, adult organism will develop. In diplohaplontic organisms a haploid phase (gametophyte) exists in the life cycle between meiosis and fertilization (e.g. higher plants, many algae and Fungi); the products of meiosis are spores that develop as haploid individuals from which haploid gametes develop to form a diploid zygote; diplohaplontic organisms show an alternation of haploid and diploid generations. In haplontic organisms meiosis occurs in the zygote, giving rise to four haploid cells (e.g. many algae and protozoa), only the zygote is diploid and this may form a resistant spore, tiding organisms over hard times.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
regulation of mitotic metaphase/anaphase transition
Any process that modulates the frequency, rate or extent of the onset of anaphase (chromosome movement) in the mitotic cell cycle.
sperm motility
Any process involved in the controlled movement of a sperm cell.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
regulation of organelle organization
Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of an organelle.
locomotion
Self-propelled movement of a cell or organism from one location to another.
positive regulation of cell cycle
Any process that activates or increases the rate or extent of progression through the cell cycle.
positive regulation of mitosis
Any process that activates or increases the frequency, rate or extent of mitosis.
positive regulation of mitotic metaphase/anaphase transition
Any process that activates or increases the frequency, rate or extent of the mitotic metaphase to anaphase transition.
male gamete generation
Generation of the male gamete; specialised haploid cells produced by meiosis and along with a female gamete takes part in sexual reproduction.
organelle fission
The creation of two or more organelles by division of one organelle.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
cell motility
Any process involved in the controlled self-propelled movement of a cell that results in translocation of the cell from one place to another.
chromosome localization
Any process by which a chromosome is transported to, or maintained in, a specific location.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cellular component organization
Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of cell structures, including the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
positive regulation of cellular component organization
Any process that activates or increases the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of cell structures, including the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
establishment of chromosome localization
The directed movement of a chromosome to a specific location.
chromosome movement towards spindle pole
The directed movement of chromosomes in the center of the spindle towards the spindle poles, mediated by the shortening of microtubules attached to the chromosomes.
meiotic cell cycle
Progression through the phases of the meiotic cell cycle, in which canonically a cell replicates to produce four offspring with half the chromosomal content of the progenitor cell.
M phase of meiotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the meiotic cell cycle during which meiosis takes place.
organelle localization
Any process by which an organelle is transported to, and/or maintained in, a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
establishment of organelle localization
The directed movement of an organelle to a specific location.
localization of cell
Any process by which a cell is transported to, and/or maintained in, a specific location.
regulation of nuclear division
Any process that modulates the frequency, rate or extent of nuclear division, the partitioning of the nucleus and its genetic information.
positive regulation of nuclear division
Any process that activates or increases the frequency, rate or extent of nuclear division, the partitioning of the nucleus and its genetic information.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
positive regulation of cell cycle process
Any process that increases the rate, frequency or extent of a cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
all
NA
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
positive regulation of cellular component organization
Any process that activates or increases the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of cell structures, including the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cellular component organization
Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of cell structures, including the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
fertilization
The union of gametes of opposite sexes during the process of sexual reproduction to form a zygote. It involves the fusion of the gametic nuclei (karyogamy) and cytoplasm (plasmogamy).
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
cell motility
Any process involved in the controlled self-propelled movement of a cell that results in translocation of the cell from one place to another.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
sister chromatid segregation
The process by which sister chromatids are organized and then physically separated and apportioned to two or more sets.
positive regulation of organelle organization
Any process that increases the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of an organelle.
positive regulation of cell cycle
Any process that activates or increases the rate or extent of progression through the cell cycle.
positive regulation of cell cycle process
Any process that increases the rate, frequency or extent of a cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
regulation of organelle organization
Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of an organelle.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
regulation of organelle organization
Any process that modulates the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of an organelle.
positive regulation of cellular component organization
Any process that activates or increases the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of cell structures, including the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
positive regulation of organelle organization
Any process that increases the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of an organelle.
gamete generation
The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell.
cell motility
Any process involved in the controlled self-propelled movement of a cell that results in translocation of the cell from one place to another.
establishment of chromosome localization
The directed movement of a chromosome to a specific location.
mitotic chromosome movement towards spindle pole
The cell cycle process whereby the directed movement of chromosomes from the center of the spindle towards the spindle poles occurs. This mediates by the shortening of microtubules attached to the chromosomes, during mitosis.
sister chromatid segregation
The process by which sister chromatids are organized and then physically separated and apportioned to two or more sets.
positive regulation of organelle organization
Any process that increases the frequency, rate or extent of a process involved in the formation, arrangement of constituent parts, or disassembly of an organelle.
regulation of mitotic cell cycle
Any process that modulates the rate or extent of progress through the mitotic cell cycle.
regulation of cell cycle process
Any process that modulates a cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
positive regulation of cell cycle
Any process that activates or increases the rate or extent of progression through the cell cycle.
positive regulation of cell cycle process
Any process that increases the rate, frequency or extent of a cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
positive regulation of mitosis
Any process that activates or increases the frequency, rate or extent of mitosis.
establishment of organelle localization
The directed movement of an organelle to a specific location.
chromosome movement towards spindle pole
The directed movement of chromosomes in the center of the spindle towards the spindle poles, mediated by the shortening of microtubules attached to the chromosomes.
mitotic chromosome movement towards spindle pole
The cell cycle process whereby the directed movement of chromosomes from the center of the spindle towards the spindle poles occurs. This mediates by the shortening of microtubules attached to the chromosomes, during mitosis.
positive regulation of mitosis
Any process that activates or increases the frequency, rate or extent of mitosis.
regulation of mitosis
Any process that modulates the frequency, rate or extent of mitosis.
positive regulation of nuclear division
Any process that activates or increases the frequency, rate or extent of nuclear division, the partitioning of the nucleus and its genetic information.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
regulation of nuclear division
Any process that modulates the frequency, rate or extent of nuclear division, the partitioning of the nucleus and its genetic information.
positive regulation of nuclear division
Any process that activates or increases the frequency, rate or extent of nuclear division, the partitioning of the nucleus and its genetic information.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
regulation of mitosis
Any process that modulates the frequency, rate or extent of mitosis.
positive regulation of mitotic metaphase/anaphase transition
Any process that activates or increases the frequency, rate or extent of the mitotic metaphase to anaphase transition.
meiosis
A cell cycle process comprising the steps by which a cell progresses through the nuclear division phase of a meiotic cell cycle, the specialized nuclear and cell division in which a single diploid cell undergoes two nuclear divisions following a single round of DNA replication in order to produce four daughter cells that contain half the number of chromosomes as the diploid cell. Meiotic division occurs during the formation of gametes from diploid organisms and at the beginning of haplophase in those organisms that alternate between diploid and haploid generations.
positive regulation of mitotic metaphase/anaphase transition
Any process that activates or increases the frequency, rate or extent of the mitotic metaphase to anaphase transition.
regulation of mitotic metaphase/anaphase transition
Any process that modulates the frequency, rate or extent of the onset of anaphase (chromosome movement) in the mitotic cell cycle.
positive regulation of mitosis
Any process that activates or increases the frequency, rate or extent of mitosis.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
M phase of meiotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the meiotic cell cycle during which meiosis takes place.
mitotic sister chromatid segregation
The cell cycle process whereby replicated homologous chromosomes are organized and then physically separated and apportioned to two sets during the mitotic cell cycle. Each replicated chromosome, composed of two sister chromatids, aligns at the cell equator, paired with its homologous partner. One homolog of each morphologic type goes into each of the resulting chromosome sets.
regulation of mitosis
Any process that modulates the frequency, rate or extent of mitosis.
mitotic metaphase/anaphase transition
The cell cycle process whereby a cell progresses from metaphase to anaphase during mitosis, triggered by the destruction of mitotic cyclins.
positive regulation of mitosis
Any process that activates or increases the frequency, rate or extent of mitosis.
establishment of chromosome localization
The directed movement of a chromosome to a specific location.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
outer dense fiber
Structure or material found in the flagella of mammalian sperm that surrounds each of the nine microtubule doublets, giving a 9 + 9 + 2 arrangement rather than the 9 + 2 pattern usually seen. These dense fibers are stiff and noncontractile.
axoneme
The bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
microtubule-based flagellum
A long, whiplike protrusion from the surface of a eukaryotic cell, whose undulations drive the cell through a liquid medium; similar in structure to a cilium. The flagellum is based on a 9+2 arrangement of microtubules.
flagellum
Long whiplike or feathery structures borne either singly or in groups by the motile cells of many bacteria and unicellular eukaryotes and by the motile male gametes of many eukaryotic organisms, which propel the cell through a liquid medium.
perinuclear theca
A condensed cytoplasmic structure that covers the nucleus of mammalian spermatozoa except for a narrow zone around the insertion of the tail. It shows two distinct regions, a subacrosomal layer and, continuing caudally beyond the acrosomic system, the postacrosomal sheath. The perinuclear theca has been considered a cytoskeletal scaffold responsible for maintaining the overall architecture of the mature sperm head; however, recent studies indicate that the bulk of its constituent proteins are not traditional cytoskeletal proteins but rather a variety of cytosolic proteins.
cytoskeletal calyx
A large cytoskeletal structure located at the posterior end of the perinuclear theca of a mammalian sperm head. The nucleus is tightly associated with the calyx, which contains calicin and basic cylicin proteins.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
axoneme part
Any constituent part of an axoneme, the bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
perinuclear region of cytoplasm
Cytoplasm situated near, or occurring around, the nucleus.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
axoneme part
Any constituent part of an axoneme, the bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
flagellum
Long whiplike or feathery structures borne either singly or in groups by the motile cells of many bacteria and unicellular eukaryotes and by the motile male gametes of many eukaryotic organisms, which propel the cell through a liquid medium.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
axoneme
The bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
axoneme part
Any constituent part of an axoneme, the bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
axoneme
The bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
axoneme part
Any constituent part of an axoneme, the bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
axoneme
The bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
microtubule-based flagellum
A long, whiplike protrusion from the surface of a eukaryotic cell, whose undulations drive the cell through a liquid medium; similar in structure to a cilium. The flagellum is based on a 9+2 arrangement of microtubules.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoskeletal calyx
A large cytoskeletal structure located at the posterior end of the perinuclear theca of a mammalian sperm head. The nucleus is tightly associated with the calyx, which contains calicin and basic cylicin proteins.
perinuclear theca
A condensed cytoplasmic structure that covers the nucleus of mammalian spermatozoa except for a narrow zone around the insertion of the tail. It shows two distinct regions, a subacrosomal layer and, continuing caudally beyond the acrosomic system, the postacrosomal sheath. The perinuclear theca has been considered a cytoskeletal scaffold responsible for maintaining the overall architecture of the mature sperm head; however, recent studies indicate that the bulk of its constituent proteins are not traditional cytoskeletal proteins but rather a variety of cytosolic proteins.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
cytoskeletal calyx
A large cytoskeletal structure located at the posterior end of the perinuclear theca of a mammalian sperm head. The nucleus is tightly associated with the calyx, which contains calicin and basic cylicin proteins.
perinuclear theca
A condensed cytoplasmic structure that covers the nucleus of mammalian spermatozoa except for a narrow zone around the insertion of the tail. It shows two distinct regions, a subacrosomal layer and, continuing caudally beyond the acrosomic system, the postacrosomal sheath. The perinuclear theca has been considered a cytoskeletal scaffold responsible for maintaining the overall architecture of the mature sperm head; however, recent studies indicate that the bulk of its constituent proteins are not traditional cytoskeletal proteins but rather a variety of cytosolic proteins.

ADAM20ADAM metallopeptidase domain 20 (ENSG00000134007), score: 0.9 AGBL3ATP/GTP binding protein-like 3 (ENSG00000146856), score: 0.91 AKNAD1AKNA domain containing 1 (ENSG00000162641), score: 0.91 ALS2CR11amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 (ENSG00000155754), score: 0.91 APLP2amyloid beta (A4) precursor-like protein 2 (ENSG00000084234), score: -0.76 APOBEC4apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) (ENSG00000173627), score: 0.91 ASB17ankyrin repeat and SOCS box-containing 17 (ENSG00000154007), score: 0.9 ASPMasp (abnormal spindle) homolog, microcephaly associated (Drosophila) (ENSG00000066279), score: 0.92 ASZ1ankyrin repeat, SAM and basic leucine zipper domain containing 1 (ENSG00000154438), score: 0.92 ATP1A4ATPase, Na+/K+ transporting, alpha 4 polypeptide (ENSG00000132681), score: 0.9 BRIP1BRCA1 interacting protein C-terminal helicase 1 (ENSG00000136492), score: 0.9 C10orf122chromosome 10 open reading frame 122 (ENSG00000175018), score: 0.92 C10orf27chromosome 10 open reading frame 27 (ENSG00000166220), score: 0.91 C10orf67chromosome 10 open reading frame 67 (ENSG00000179133), score: 0.91 C10orf96chromosome 10 open reading frame 96 (ENSG00000182645), score: 0.91 C12orf56chromosome 12 open reading frame 56 (ENSG00000185306), score: 0.92 C12orf63chromosome 12 open reading frame 63 (ENSG00000188596), score: 0.93 C12orf71chromosome 12 open reading frame 71 (ENSG00000214700), score: 0.93 C14orf145chromosome 14 open reading frame 145 (ENSG00000100629), score: 0.92 C14orf39chromosome 14 open reading frame 39 (ENSG00000179008), score: 0.98 C16orf78chromosome 16 open reading frame 78 (ENSG00000166152), score: 0.91 C17orf46chromosome 17 open reading frame 46 (ENSG00000184361), score: 0.9 C18orf34chromosome 18 open reading frame 34 (ENSG00000166960), score: 0.92 C1orf100chromosome 1 open reading frame 100 (ENSG00000173728), score: 0.91 C1orf101chromosome 1 open reading frame 101 (ENSG00000179397), score: 0.9 C1orf129chromosome 1 open reading frame 129 (ENSG00000117501), score: 0.93 C1orf141chromosome 1 open reading frame 141 (ENSG00000203963), score: 0.96 C1orf185chromosome 1 open reading frame 185 (ENSG00000204006), score: 0.94 C1orf65chromosome 1 open reading frame 65 (ENSG00000178395), score: 0.9 C20orf173chromosome 20 open reading frame 173 (ENSG00000125975), score: 0.92 C3orf22chromosome 3 open reading frame 22 (ENSG00000180697), score: 0.91 C3orf77chromosome 3 open reading frame 77 (ENSG00000173769), score: 0.91 C4orf17chromosome 4 open reading frame 17 (ENSG00000138813), score: 0.91 C5orf48chromosome 5 open reading frame 48 (ENSG00000196900), score: 0.96 C5orf50chromosome 5 open reading frame 50 (ENSG00000185662), score: 0.91 C6orf103chromosome 6 open reading frame 103 (ENSG00000118492), score: 0.91 C7orf57chromosome 7 open reading frame 57 (ENSG00000164746), score: 0.94 C9orf93chromosome 9 open reading frame 93 (ENSG00000164989), score: 0.91 CAPN1calpain 1, (mu/I) large subunit (ENSG00000014216), score: -0.78 CCDC15coiled-coil domain containing 15 (ENSG00000149548), score: 0.93 CCDC22coiled-coil domain containing 22 (ENSG00000101997), score: -0.83 CCDC38coiled-coil domain containing 38 (ENSG00000165972), score: 0.96 CCDC67coiled-coil domain containing 67 (ENSG00000165325), score: 0.94 CCDC83coiled-coil domain containing 83 (ENSG00000150676), score: 0.93 CDCA7cell division cycle associated 7 (ENSG00000144354), score: 0.91 CDKL3cyclin-dependent kinase-like 3 (ENSG00000006837), score: 0.9 CENPEcentromere protein E, 312kDa (ENSG00000138778), score: 0.94 CEP55centrosomal protein 55kDa (ENSG00000138180), score: 0.91 CKAP2Lcytoskeleton associated protein 2-like (ENSG00000169607), score: 0.9 CLCA2chloride channel accessory 2 (ENSG00000137975), score: 0.97 CNBD1cyclic nucleotide binding domain containing 1 (ENSG00000176571), score: 0.98 CPXCR1CPX chromosome region, candidate 1 (ENSG00000147183), score: 0.92 CST8cystatin 8 (cystatin-related epididymal specific) (ENSG00000125815), score: 0.92 CSTL1cystatin-like 1 (ENSG00000125823), score: 0.98 CTBP1C-terminal binding protein 1 (ENSG00000159692), score: -0.75 CXorf22chromosome X open reading frame 22 (ENSG00000165164), score: 0.95 CXorf30chromosome X open reading frame 30 (ENSG00000205081), score: 0.93 CYLC1cylicin, basic protein of sperm head cytoskeleton 1 (ENSG00000183035), score: 0.91 CYLC2cylicin, basic protein of sperm head cytoskeleton 2 (ENSG00000155833), score: 0.92 CYP19A1cytochrome P450, family 19, subfamily A, polypeptide 1 (ENSG00000137869), score: 0.92 DLGAP5discs, large (Drosophila) homolog-associated protein 5 (ENSG00000126787), score: 0.94 DNAH8dynein, axonemal, heavy chain 8 (ENSG00000124721), score: 0.93 DNAJB7DnaJ (Hsp40) homolog, subfamily B, member 7 (ENSG00000172404), score: 0.93 DYDC1DPY30 domain containing 1 (ENSG00000170788), score: 0.9 E2F2E2F transcription factor 2 (ENSG00000007968), score: 0.96 E2F7E2F transcription factor 7 (ENSG00000165891), score: 0.91 EFCAB5EF-hand calcium binding domain 5 (ENSG00000176927), score: 0.93 EFHBEF-hand domain family, member B (ENSG00000163576), score: 0.92 ENTHD1ENTH domain containing 1 (ENSG00000176177), score: 0.93 ESCO2establishment of cohesion 1 homolog 2 (S. cerevisiae) (ENSG00000171320), score: 0.94 EXD1exonuclease 3'-5' domain containing 1 (ENSG00000178997), score: 0.91 FAM194Afamily with sequence similarity 194, member A (ENSG00000163645), score: 0.9 FAM46Dfamily with sequence similarity 46, member D (ENSG00000174016), score: 0.95 FAM71Bfamily with sequence similarity 71, member B (ENSG00000170613), score: 0.91 FAM71Dfamily with sequence similarity 71, member D (ENSG00000172717), score: 0.92 FBXO43F-box protein 43 (ENSG00000156509), score: 0.92 FBXO47F-box protein 47 (ENSG00000204952), score: 0.91 FCRL4Fc receptor-like 4 (ENSG00000163518), score: 0.96 FLOT2flotillin 2 (ENSG00000132589), score: -0.77 GPR18G protein-coupled receptor 18 (ENSG00000125245), score: 0.93 GSG2germ cell associated 2 (haspin) (ENSG00000177602), score: 0.91 H1FOOH1 histone family, member O, oocyte-specific (ENSG00000178804), score: 0.95 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (ENSG00000126945), score: -0.76 HORMAD1HORMA domain containing 1 (ENSG00000143452), score: 0.92 HOXA13homeobox A13 (ENSG00000106031), score: 1 IFLTD1intermediate filament tail domain containing 1 (ENSG00000152936), score: 0.96 IGF2BP3insulin-like growth factor 2 mRNA binding protein 3 (ENSG00000136231), score: 0.9 IQCHIQ motif containing H (ENSG00000103599), score: 0.91 JDP2Jun dimerization protein 2 (ENSG00000140044), score: -0.79 KCNU1potassium channel, subfamily U, member 1 (ENSG00000215262), score: 0.94 KIF14kinesin family member 14 (ENSG00000118193), score: 0.92 KIF18Akinesin family member 18A (ENSG00000121621), score: 0.92 LASS3LAG1 homolog, ceramide synthase 3 (ENSG00000154227), score: 0.91 LCTlactase (ENSG00000115850), score: 0.91 LEKR1leucine, glutamate and lysine rich 1 (ENSG00000197980), score: 0.96 LRGUKleucine-rich repeats and guanylate kinase domain containing (ENSG00000155530), score: 0.92 LRRC63leucine rich repeat containing 63 (ENSG00000173988), score: 0.9 LRRC67leucine rich repeat containing 67 (ENSG00000178125), score: 0.93 LRRIQ4leucine-rich repeats and IQ motif containing 4 (ENSG00000188306), score: 0.96 MAGEA8melanoma antigen family A, 8 (ENSG00000156009), score: 0.91 MBD3L1methyl-CpG binding domain protein 3-like 1 (ENSG00000170948), score: 0.9 MOSv-mos Moloney murine sarcoma viral oncogene homolog (ENSG00000172680), score: 0.94 MPP1membrane protein, palmitoylated 1, 55kDa (ENSG00000130830), score: -0.76 MS4A5membrane-spanning 4-domains, subfamily A, member 5 (ENSG00000166930), score: 0.93 MYO1Amyosin IA (ENSG00000166866), score: 0.96 NAA10N(alpha)-acetyltransferase 10, NatA catalytic subunit (ENSG00000102030), score: -0.8 NFIBnuclear factor I/B (ENSG00000147862), score: -0.76 ODF3outer dense fiber of sperm tails 3 (ENSG00000177947), score: 0.9 ODF4outer dense fiber of sperm tails 4 (ENSG00000184650), score: 0.93 PAGE3P antigen family, member 3 (prostate associated) (ENSG00000204279), score: 0.91 PDCL2phosducin-like 2 (ENSG00000163440), score: 0.91 PLAC1Lplacenta-specific 1-like (ENSG00000149507), score: 0.93 PLCZ1phospholipase C, zeta 1 (ENSG00000139151), score: 0.93 PLSCR2phospholipid scramblase 2 (ENSG00000163746), score: 0.91 PMFBP1polyamine modulated factor 1 binding protein 1 (ENSG00000118557), score: 0.9 POLQpolymerase (DNA directed), theta (ENSG00000051341), score: 0.96 PQBP1polyglutamine binding protein 1 (ENSG00000102103), score: -0.85 PRDX2peroxiredoxin 2 (ENSG00000167815), score: -0.84 PRM3protamine 3 (ENSG00000178257), score: 0.91 PRSS37protease, serine, 37 (ENSG00000165076), score: 0.91 RAD51AP1RAD51 associated protein 1 (ENSG00000111247), score: 0.93 RNF113Aring finger protein 113A (ENSG00000125352), score: -0.77 RNF17ring finger protein 17 (ENSG00000132972), score: 0.91 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (ENSG00000092200), score: 0.91 SCELsciellin (ENSG00000136155), score: 0.91 SEL1L2sel-1 suppressor of lin-12-like 2 (C. elegans) (ENSG00000101251), score: 0.92 SEPT14septin 14 (ENSG00000154997), score: 0.9 SERPINB5serpin peptidase inhibitor, clade B (ovalbumin), member 5 (ENSG00000206075), score: 0.93 SGOL2shugoshin-like 2 (S. pombe) (ENSG00000163535), score: 0.93 SLC9A10solute carrier family 9, member 10 (ENSG00000172139), score: 0.97 SLC9A11solute carrier family 9, member 11 (ENSG00000162753), score: 0.97 SLCO6A1solute carrier organic anion transporter family, member 6A1 (ENSG00000205359), score: 0.92 SMAD7SMAD family member 7 (ENSG00000101665), score: -0.74 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (ENSG00000106304), score: 0.96 SPATA9spermatogenesis associated 9 (ENSG00000145757), score: 0.9 SPC25SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000152253), score: 0.94 STARD6StAR-related lipid transfer (START) domain containing 6 (ENSG00000174448), score: 0.91 SYCP1synaptonemal complex protein 1 (ENSG00000198765), score: 0.93 TEX15testis expressed 15 (ENSG00000133863), score: 0.9 TFAP2Dtranscription factor AP-2 delta (activating enhancer binding protein 2 delta) (ENSG00000008197), score: 0.95 TIGD4tigger transposable element derived 4 (ENSG00000169989), score: 0.9 TMC5transmembrane channel-like 5 (ENSG00000103534), score: 0.93 TMEM146transmembrane protein 146 (ENSG00000174898), score: 0.93 TMEM225transmembrane protein 225 (ENSG00000204300), score: 0.9 TNP2transition protein 2 (during histone to protamine replacement) (ENSG00000178279), score: 0.97 TRIML1tripartite motif family-like 1 (ENSG00000184108), score: 0.92 TRIML2tripartite motif family-like 2 (ENSG00000179046), score: 0.9 TRYX3trypsin X3 (ENSG00000171147), score: 0.92 UBL4Aubiquitin-like 4A (ENSG00000102178), score: -0.77 USP26ubiquitin specific peptidase 26 (ENSG00000134588), score: 0.94 VASH2vasohibin 2 (ENSG00000143494), score: 0.9 WDR45WD repeat domain 45 (ENSG00000196998), score: -0.87 WDR64WD repeat domain 64 (ENSG00000162843), score: 0.95 XAGE5X antigen family, member 5 (ENSG00000171405), score: 0.96 ZNF358zinc finger protein 358 (ENSG00000198816), score: -0.81 ZNF572zinc finger protein 572 (ENSG00000180938), score: 0.9 ZSWIM2zinc finger, SWIM-type containing 2 (ENSG00000163012), score: 0.93
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_ts_m_ca1 | ggo | ts | m | _ |
| ppa_ts_m_ca1 | ppa | ts | m | _ |
| ptr_ts_m_ca1 | ptr | ts | m | _ |
| mml_ts_m2_ca1 | mml | ts | m | 2 |
| mml_ts_m1_ca1 | mml | ts | m | 1 |