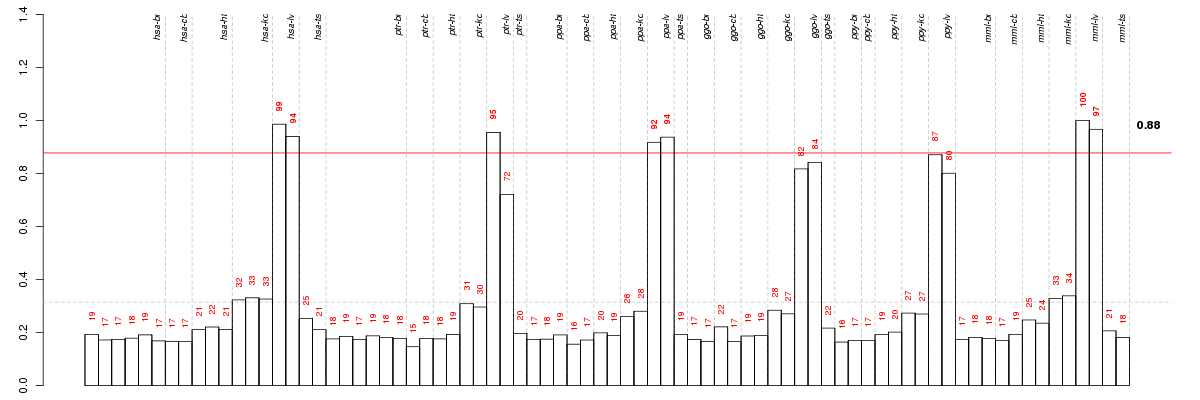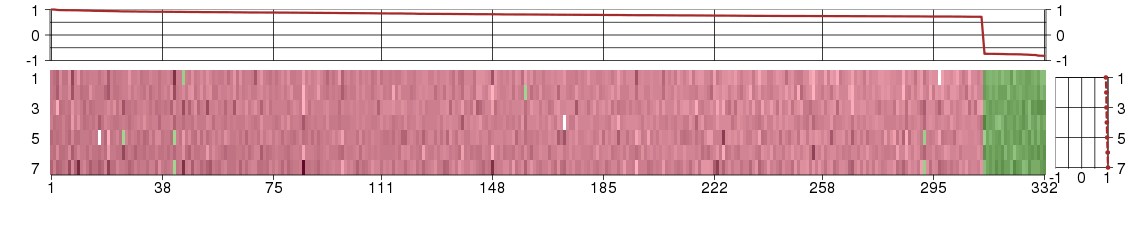

Under-expression is coded with green,
over-expression with red color.

sulfur amino acid metabolic process
The chemical reactions and pathways involving amino acids containing sulfur, comprising cysteine, homocysteine, methionine and selenocysteine.
sulfur metabolic process
The chemical reactions and pathways involving the nonmetallic element sulfur or compounds that contain sulfur, such as the amino acids methionine and cysteine or the tripeptide glutathione.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
adaptive immune response
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process, and allowing for enhanced response to subsequent exposures to the same antigen (immunological memory).
immune effector process
Any process of the immune system that occurs as part of an immune response.
activation of immune response
Any process that initiates an immune response.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
leukocyte mediated immunity
Any process involved in the carrying out of an immune response by a leukocyte.
lymphocyte mediated immunity
Any process involved in the carrying out of an immune response by a lymphocyte.
humoral immune response mediated by circulating immunoglobulin
An immune response dependent upon secreted immunoglobulin. An example of this process is found in Mus musculus.
adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process that includes somatic recombination of germline gene segments encoding immunoglobulin superfamily domains, and allowing for enhanced responses upon subsequent exposures to the same antigen (immunological memory). Recombined receptors for antigen encoded by immunoglobulin superfamily domains include T cell receptors and immunoglobulins (antibodies). An example of this is the adaptive immune response found in Mus musculus.
acute inflammatory response
Inflammation which comprises a rapid, short-lived, relatively uniform response to acute injury or antigenic challenge and is characterized by accumulations of fluid, plasma proteins, and granulocytic leukocytes. An acute inflammatory response occurs within a matter of minutes or hours, and either resolves within a few days or becomes a chronic inflammatory response.
activation of plasma proteins involved in acute inflammatory response
Any process activating plasma proteins by proteolysis as part of an acute inflammatory response.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
acute-phase response
Process involving non-antibody proteins whose concentrations in the plasma increase in response to infection or injury of homeothermic animals.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
carbohydrate metabolic process
The chemical reactions and pathways involving carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y. Includes the formation of carbohydrate derivatives by the addition of a carbohydrate residue to another molecule.
polysaccharide metabolic process
The chemical reactions and pathways involving polysaccharides, a polymer of more than 20 monosaccharide residues joined by glycosidic linkages.
monosaccharide metabolic process
The chemical reactions and pathways involving monosaccharides, the simplest carbohydrates. They are polyhydric alcohols containing either an aldehyde or a keto group and between three to ten or more carbon atoms. They form the constitutional repeating units of oligo- and polysaccharides.
glucose metabolic process
The chemical reactions and pathways involving glucose, the aldohexose gluco-hexose. D-glucose is dextrorotatory and is sometimes known as dextrose; it is an important source of energy for living organisms and is found free as well as combined in homo- and hetero-oligosaccharides and polysaccharides.
alcohol metabolic process
The chemical reactions and pathways involving alcohols, any of a class of compounds containing one or more hydroxyl groups attached to a saturated carbon atom.
cellular aldehyde metabolic process
The chemical reactions and pathways involving aldehydes, any organic compound with the formula R-CH=O, as carried out by individual cells.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
pyruvate metabolic process
The chemical reactions and pathways involving pyruvate, 2-oxopropanoate.
gluconeogenesis
The formation of glucose from noncarbohydrate precursors, such as pyruvate, amino acids and glycerol.
oxidation reduction
The process of removal or addition of one or more electrons with or without the concomitant removal or addition of a proton or protons.
protein maturation by peptide bond cleavage
The hydrolysis of a peptide bond or bonds within a protein as part of protein maturation, the process leading to the attainment of the full functional capacity of a protein.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cysteine metabolic process
The chemical reactions and pathways involving cysteine, 2-amino-3-mercaptopropanoic acid.
glycine metabolic process
The chemical reactions and pathways involving glycine, aminoethanoic acid.
histidine metabolic process
The chemical reactions and pathways involving histidine, 2-amino-3-(1H-imidazol-4-yl)propanoic acid.
histidine catabolic process
The chemical reactions and pathways resulting in the breakdown of histidine, 2-amino-3-(1H-imidazol-4-yl)propanoic acid.
cellular amino acid derivative metabolic process
The chemical reactions and pathways involving compounds derived from amino acids, organic acids containing one or more amino substituents.
cellular biogenic amine metabolic process
The chemical reactions and pathways occurring at the level of individual cells involving any of a group of naturally occurring, biologically active amines, such as norepinephrine, histamine, and serotonin, many of which act as neurotransmitters.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
neutral lipid metabolic process
The chemical reactions and pathways involving neutral lipids, lipids only soluble in solvents of very low polarity.
acylglycerol metabolic process
The chemical reactions and pathways involving acylglycerol, any mono-, di- or triester of glycerol with (one or more) fatty acids.
triglyceride metabolic process
The chemical reactions and pathways involving triglyceride, any triester of glycerol. The three fatty acid residues may all be the same or differ in any permutation. Triglycerides are important components of plant oils, animal fats and animal plasma lipoproteins.
phospholipid metabolic process
The chemical reactions and pathways involving phospholipids, any lipid containing phosphoric acid as a mono- or diester.
glycerophospholipid metabolic process
The chemical reactions and pathways involving glycerophospholipids, any derivative of glycerophosphate that contains at least one O-acyl, O-alkyl, or O-alkenyl group attached to the glycerol residue.
glycerol ether metabolic process
The chemical reactions and pathways involving glycerol ethers, any anhydride formed between two organic hydroxy compounds, one of which is glycerol.
steroid biosynthetic process
The chemical reactions and pathways resulting in the formation of steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus; includes de novo formation and steroid interconversion by modification.
cholesterol biosynthetic process
The chemical reactions and pathways resulting in the formation of cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones.
steroid catabolic process
The chemical reactions and pathways resulting in the breakdown of steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
cellular aromatic compound metabolic process
The chemical reactions and pathways involving aromatic compounds, any organic compound characterized by one or more planar rings, each of which contains conjugated double bonds and delocalized pi electrons, as carried out by individual cells.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
lipid transport
The directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore. Lipids are compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
complement activation, alternative pathway
Any process involved in the activation of any of the steps of the alternative pathway of the complement cascade which allows for the direct killing of microbes and the regulation of other immune processes.
complement activation, classical pathway
Any process involved in the activation of any of the steps of the classical pathway of the complement cascade which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes.
humoral immune response
An immune response mediated through a body fluid.
response to nutrient
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
blood coagulation, extrinsic pathway
A pathway of blood coagulation in which the earlier stages of the cascade are bypassed and the activation of factor X to factor Xa is effected by the combination of factor VIIa + thromboplastin; this second pathway occurs when tissue extracts are present in optimal amounts and is much more rapid than the intrinsic pathway.
hemostasis
The stopping of bleeding (loss of body fluid) or the arrest of the circulation to an organ or part.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
cholesterol metabolic process
The chemical reactions and pathways involving cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones. It is a component of the plasma membrane lipid bilayer and of plasma lipoproteins and can be found in all animal tissues.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
cell death
A biological process that results in permanent cessation of all vital functions of a cell.
cytolysis
The rupture of cell membranes and the loss of cytoplasm.
negative regulation of coagulation
Any process that stops, prevents or reduces the frequency, rate or extent of coagulation.
lipid biosynthetic process
The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
cellular amino acid biosynthetic process
The chemical reactions and pathways resulting in the formation of amino acids, organic acids containing one or more amino substituents.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
cellular amino acid catabolic process
The chemical reactions and pathways resulting in the breakdown of amino acids, organic acids containing one or more amino substituents.
serine family amino acid metabolic process
The chemical reactions and pathways involving amino acids of the serine family, comprising cysteine, glycine, homoserine, selenocysteine and serine.
histidine family amino acid metabolic process
The chemical reactions and pathways involving amino acids of the histidine family.
histidine family amino acid catabolic process
The chemical reactions and pathways resulting in the breakdown of amino acids of the histidine family.
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
amine biosynthetic process
The chemical reactions and pathways resulting in the formation of any organic compound that is weakly basic in character and contains an amino or a substituted amino group. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
amine catabolic process
The chemical reactions and pathways resulting in the breakdown of any organic compound that is weakly basic in character and contains an amino or a substituted amino group. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
response to drug
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a drug stimulus. A drug is a substance used in the diagnosis, treatment or prevention of a disease.
response to xenobiotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a xenobiotic compound stimulus. Xenobiotic compounds are compounds foreign to living organisms.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
response to endogenous stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus arising within the organism.
response to hormone stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hormone stimulus.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to extracellular stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
response to organic substance
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic substance stimulus.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of hormone levels
Any process that modulates the levels of hormone within an organism or a tissue. A hormone is any substance formed in very small amounts in one specialized organ or group of cells and carried (sometimes in the bloodstream) to another organ or group of cells in the same organism, upon which it has a specific regulatory action.
lipid localization
Any process by which a lipid is transported to, or maintained in, a specific location.
cellular component assembly
The aggregation, arrangement and bonding together of a cellular component.
sterol transport
The directed movement of sterols into, out of, within or between cells by means of some external agent such as a transporter or pore. Sterols are steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y.
organic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of organic acids, any acidic compound containing carbon in covalent linkage.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
immunoglobulin mediated immune response
An immune response mediated by immunoglobulins, whether cell-bound or in solution.
sterol metabolic process
The chemical reactions and pathways involving sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
sterol biosynthetic process
The chemical reactions and pathways resulting in the formation of sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
death
A permanent cessation of all vital functions: the end of life; can be applied to a whole organism or to a part of an organism.
protein processing
Any protein maturation process achieved by the cleavage of peptide bonds.
drug metabolic process
The chemical reactions and pathways involving a drug, a substance used in the diagnosis, treatment or prevention of a disease; as used here antibiotic substances (see antibiotic metabolism) are considered to be drugs, even if not used in medical or veterinary practice.
organic ether metabolic process
The chemical reactions and pathways involving organic ethers, any anhydride of the general formula R1-O-R2, formed between two identical or nonidentical organic hydroxy compounds.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
hexose metabolic process
The chemical reactions and pathways involving a hexose, any monosaccharide with a chain of six carbon atoms in the molecule.
hexose biosynthetic process
The chemical reactions and pathways resulting in the formation of hexose, any monosaccharide with a chain of six carbon atoms in the molecule.
aromatic compound catabolic process
The chemical reactions and pathways resulting in the breakdown of aromatic compounds, any substance containing an aromatic carbon ring.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
organophosphate metabolic process
The chemical reactions and pathways involving organophosphates, any phosphate-containing organic compound.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
secondary metabolic process
The chemical reactions and pathways resulting in many of the chemical changes of compounds that are not necessarily required for growth and maintenance of cells, and are often unique to a taxon. In multicellular organisms secondary metabolism is generally carried out in specific cell types, and may be useful for the organism as a whole. In unicellular organisms, secondary metabolism is often used for the production of antibiotics or for the utilization and acquisition of unusual nutrients.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
regulation of blood coagulation
Any process that modulates the frequency, rate or extent of blood coagulation.
positive regulation of blood coagulation
Any process that activates or increases the frequency, rate or extent of blood coagulation.
negative regulation of blood coagulation
Any process that stops, prevents or reduces the frequency, rate or extent of blood coagulation.
cholesterol transport
The directed movement of cholesterol, cholest-5-en-3-beta-ol, into, out of, within or between cells by means of some external agent such as a transporter or pore.
bile acid catabolic process
The chemical reactions and pathways resulting in the breakdown of bile acids, any of a group of steroid carboxylic acids occurring in bile.
response to nutrient levels
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients.
regulation of response to external stimulus
Any process that modulates the frequency, rate or extent of a response to an external stimulus.
regulation of lipid transport
Any process that modulates the frequency, rate or extent of the directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore.
negative regulation of lipid transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore.
regulation of sterol transport
Any process that modulates the frequency, rate or extent of the directed movement of sterols into, out of, within or between cells by means of some external agent such as a transporter or pore.
negative regulation of sterol transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of sterols into, out of, within or between cells by means of some external agent such as a transporter or pore.
regulation of cholesterol transport
Any process that modulates the frequency, rate or extent of the directed movement of cholesterol into, out of, within or between cells by means of some external agent such as a transporter or pore.
negative regulation of cholesterol transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of cholesterol into, out of, within or between cells by means of some external agent such as a transporter or pore.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
monocarboxylic acid metabolic process
The chemical reactions and pathways involving monocarboxylic acids, any organic acid containing one carboxyl (COOH) group or anion (COO-).
regulation of localization
Any process that modulates the frequency, rate or extent of any process by which a cell, a substance, or a cellular entity is transported to, or maintained in, a specific location.
macromolecule localization
Any process by which a macromolecule is transported to, or maintained in, a specific location.
cholesterol efflux
The directed movement of cholesterol, cholest-5-en-3-beta-ol, out of a cell or organelle.
macromolecular complex remodeling
The acquisition, loss, or modification of macromolecules within a complex, resulting in the alteration of an existing complex.
protein-lipid complex remodeling
The acquisition, loss or modification of a protein or lipid within a protein-lipid complex.
plasma lipoprotein particle remodeling
The acquisition, loss or modification of a protein or lipid within a plasma lipoprotein particle, including the hydrolysis of triglyceride by hepatic lipase, with the subsequent loss of free fatty acid, and the esterification of cholesterol by phosphatidylcholine-sterol O-acyltransferase (lecithin cholesterol acyltransferase; LCAT).
triglyceride-rich lipoprotein particle remodeling
The acquisition, loss or modification of a protein or lipid within a triglyceride-rich lipoprotein particle, including the hydrolysis of triglyceride by lipoprotein lipase, with the subsequent loss of free fatty acid, and the transfer of cholesterol esters to a triglyceride-rich lipoprotein particle by cholesteryl ester transfer protein (CETP), with the simultaneous transfer of triglyceride from a triglyceride-rich lipoprotein particle.
very-low-density lipoprotein particle remodeling
The acquisition, loss or modification of a protein or lipid within a very-low-density lipoprotein particle, including the hydrolysis of triglyceride by hepatic lipase or lipoprotein lipase and the subsequent loss of free fatty acid.
low-density lipoprotein particle remodeling
The acquisition, loss or modification of a protein or lipid within a low-density lipoprotein particle, including the hydrolysis of triglyceride by hepatic lipase, with the subsequent loss of free fatty acid, and the transfer of cholesterol esters from LDL to a triglyceride-rich lipoprotein particle by cholesteryl ester transfer protein (CETP), with the simultaneous transfer of triglyceride to LDL.
high-density lipoprotein particle remodeling
The acquisition, loss or modification of a protein or lipid within a high-density lipoprotein particle, including the hydrolysis of triglyceride by hepatic lipase, with the subsequent loss of free fatty acid, and the transfer of cholesterol esters from LDL to a triglyceride-rich lipoprotein particle by cholesteryl ester transfer protein (CETP), with the simultaneous transfer of triglyceride to LDL.
plasma lipoprotein particle assembly
The aggregation and arrangement of proteins and lipids to form a plasma lipoprotein particle.
very-low-density lipoprotein particle assembly
The aggregation and arrangement of proteins and lipids in the liver to form a very-low-density lipoprotein particle.
lipoprotein particle clearance
The process by which a lipoprotein particle is removed from the blood via receptor-mediated endocytosis and its constituent parts degraded.
chylomicron remnant clearance
The process by which a chylomicron remnant is removed from the blood via receptor-mediated endocytosis into liver cells and its constituent parts degraded.
cellular carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, carried out by individual cells.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular hormone metabolic process
The chemical reactions and pathways involving any hormone, naturally occurring substances secreted by specialized cells that affects the metabolism or behavior of other cells possessing functional receptors for the hormone, as carried out by individual cells.
wound healing
The series of events that restore integrity to a damaged tissue, following an injury.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
response to chemical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
ethanolamine and derivative metabolic process
The chemical reactions and pathways involving ethanolamine (2-aminoethanol) and compounds derived from it.
hormone metabolic process
The chemical reactions and pathways involving any hormone, naturally occurring substances secreted by specialized cells that affects the metabolism or behavior of other cells possessing functional receptors for the hormone.
homeostatic process
Any biological process involved in the maintenance of an internal steady-state.
cholesterol homeostasis
Any process involved in the maintenance of an internal steady-state of cholesterol within an organism or cell.
fibrinolysis
An ongoing process that solubilizes fibrin, chiefly by the proteolytic action of plasmin, resulting in the removal of small blood clots.
drug catabolic process
The chemical reactions and pathways resulting in the breakdown of a drug, a substance used in the diagnosis, treatment or prevention of a disease.
exogenous drug catabolic process
The chemical reactions and pathways resulting in the breakdown of a drug that has originated externally to the cell or organism.
negative regulation of catalytic activity
Any process that stops or reduces the activity of an enzyme.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
response to peptide hormone stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a peptide hormone stimulus. A peptide hormone is any of a class of peptides that are secreted into the blood stream and have endocrine functions in living animals.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
macromolecular complex subunit organization
Any process by which macromolecules aggregate, disaggregate, or are modified, resulting in the formation, disassembly, or alteration of a macromolecular complex.
cellular component biogenesis
The process by which a cellular component is synthesized, aggregates, and bonds together. Includes biosynthesis of constituent macromolecules, and those macromolecular modifications that are involved in synthesis or assembly of the cellular component.
negative regulation of molecular function
Any process that stops or reduces the rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
cellular carbohydrate metabolic process
The chemical reactions and pathways involving carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, as carried out by individual cells.
cellular nitrogen compound biosynthetic process
The chemical reactions and pathways resulting in the formation of organic and inorganic nitrogenous compounds.
sulfur compound biosynthetic process
The chemical reactions and pathways resulting in the formation of compounds that contain sulfur, such as the amino acids methionine and cysteine or the tripeptide glutathione.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
small molecule biosynthetic process
The chemical reactions and pathways resulting in the formation of small molecules, any monomeric molecule of small relative molecular mass.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
alcohol biosynthetic process
The chemical reactions and pathways resulting in the formation of alcohols, any of a class of compounds containing one or more hydroxyl groups attached to a saturated carbon atom.
monosaccharide biosynthetic process
The chemical reactions and pathways resulting in the formation of monosaccharides, polyhydric alcohols containing either an aldehyde or a keto group and between three to ten or more carbon atoms.
carboxylic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
phosphatidylcholine metabolic process
The chemical reactions and pathways involving phosphatidylcholines, any of a class of glycerophospholipids in which the phosphatidyl group is esterified to the hydroxyl group of choline. They are important constituents of cell membranes.
heterocycle metabolic process
The chemical reactions and pathways involving heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
glycerolipid metabolic process
The chemical reactions and pathways involving glycerolipids, any lipid with a glycerol backbone. Diacylglycerol and phosphatidate are key lipid intermediates of glycerolipid biosynthesis.
glyoxylate metabolic process
The chemical reactions and pathways involving glyoxylate, the anion of glyoxylic acid, HOC-COOH.
glycerolipid catabolic process
The chemical reactions and pathways resulting in the breakdown of glycerolipids, any lipid with a glycerol backbone.
heterocycle catabolic process
The chemical reactions and pathways resulting in the breakdown of heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
response to steroid hormone stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a steroid hormone stimulus.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
chemical homeostasis
Any biological process involved in the maintenance of an internal steady-state of a chemical.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
coagulation
The process by which a fluid solution, or part of it, changes into a solid or semisolid mass.
regulation of coagulation
Any process that modulates the frequency, rate or extent of coagulation, the process by which a fluid solution, or part of it, changes into a solid or semisolid mass.
positive regulation of coagulation
Any process that activates or increases the frequency, rate or extent of coagulation.
regulation of body fluid levels
Any process that modulates the levels of body fluids.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of lipoprotein lipase activity
Any process that modulates the activity of the enzyme lipoprotein lipase.
negative regulation of lipoprotein lipase activity
Any process that stops or reduces the activity of the enzyme lipoprotein lipase.
regulation of transport
Any process that modulates the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells by means of some external agent such as a transporter or pore.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells by means of some external agent such as a transporter or pore.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
cofactor metabolic process
The chemical reactions and pathways involving a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
positive regulation of multicellular organismal process
Any process that activates or increases the frequency, rate or extent of an organismal process, any of the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
negative regulation of multicellular organismal process
Any process that stops, prevents or reduces the frequency, rate or extent of an organismal process, the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of hydrolase activity
Any process that modulates the frequency, rate or extent of hydrolase activity, the catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
negative regulation of hydrolase activity
Any process that stops or reduces the rate of hydrolase activity, the catalysis of the hydrolysis of various bonds.
protein maturation
Any process leading to the attainment of the full functional capacity of a protein.
regulation of fibrinolysis
Any process that modulates the frequency, rate or extent of fibrinolysis, an ongoing process that solubilizes fibrin, resulting in the removal of small blood clots.
positive regulation of fibrinolysis
Any process that activates, maintains or increases the frequency, rate or extent of fibrinolysis, an ongoing process that solubilizes fibrin, resulting in the removal of small blood clots.
lipid homeostasis
Any process involved in the maintenance of an internal steady-state of lipid within an organism or cell.
sterol homeostasis
Any process involved in the maintenance of an internal steady-state of sterol within an organism or cell.
regulation of lipase activity
Any process that modulates the frequency, rate or extent of lipase activity, the hydrolysis of a lipid or phospholipid.
negative regulation of lipase activity
Any process that decreases the frequency, rate or extent of lipase activity, the hydrolysis of a lipid or phospholipid.
regulation of wound healing
Any process that modulates the rate, frequency, or extent of the series of events that restore integrity to a damaged tissue, following an injury.
macromolecular complex assembly
The aggregation, arrangement and bonding together of a set of macromolecules to form a complex.
protein-lipid complex assembly
The aggregation, arrangement and bonding together of proteins and lipids to form a protein-lipid complex.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
regulation of molecular function
Any process that modulates the frequency, rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
triglyceride homeostasis
Any process involved in the maintenance of an internal steady-state of triglyceride within an organism or cell.
regulation of response to stress
Any process that modulates the frequency, rate or extent of a response to stress. Response to stress is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cell death
A biological process that results in permanent cessation of all vital functions of a cell.
cellular component assembly
The aggregation, arrangement and bonding together of a cellular component.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
positive regulation of multicellular organismal process
Any process that activates or increases the frequency, rate or extent of an organismal process, any of the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
negative regulation of multicellular organismal process
Any process that stops, prevents or reduces the frequency, rate or extent of an organismal process, the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of localization
Any process that modulates the frequency, rate or extent of any process by which a cell, a substance, or a cellular entity is transported to, or maintained in, a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular hormone metabolic process
The chemical reactions and pathways involving any hormone, naturally occurring substances secreted by specialized cells that affects the metabolism or behavior of other cells possessing functional receptors for the hormone, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
small molecule biosynthetic process
The chemical reactions and pathways resulting in the formation of small molecules, any monomeric molecule of small relative molecular mass.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
immune effector process
Any process of the immune system that occurs as part of an immune response.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
macromolecular complex assembly
The aggregation, arrangement and bonding together of a set of macromolecules to form a complex.
regulation of coagulation
Any process that modulates the frequency, rate or extent of coagulation, the process by which a fluid solution, or part of it, changes into a solid or semisolid mass.
positive regulation of multicellular organismal process
Any process that activates or increases the frequency, rate or extent of an organismal process, any of the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
negative regulation of multicellular organismal process
Any process that stops, prevents or reduces the frequency, rate or extent of an organismal process, the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
positive regulation of coagulation
Any process that activates or increases the frequency, rate or extent of coagulation.
negative regulation of coagulation
Any process that stops, prevents or reduces the frequency, rate or extent of coagulation.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of fibrinolysis
Any process that activates, maintains or increases the frequency, rate or extent of fibrinolysis, an ongoing process that solubilizes fibrin, resulting in the removal of small blood clots.
regulation of response to stress
Any process that modulates the frequency, rate or extent of a response to stress. Response to stress is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
regulation of response to external stimulus
Any process that modulates the frequency, rate or extent of a response to an external stimulus.
regulation of transport
Any process that modulates the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells by means of some external agent such as a transporter or pore.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells by means of some external agent such as a transporter or pore.
regulation of body fluid levels
Any process that modulates the levels of body fluids.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, as carried out by individual cells.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
cellular carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, carried out by individual cells.
cellular nitrogen compound biosynthetic process
The chemical reactions and pathways resulting in the formation of organic and inorganic nitrogenous compounds.
organic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of organic acids, any acidic compound containing carbon in covalent linkage.
protein maturation
Any process leading to the attainment of the full functional capacity of a protein.
sulfur compound biosynthetic process
The chemical reactions and pathways resulting in the formation of compounds that contain sulfur, such as the amino acids methionine and cysteine or the tripeptide glutathione.
organic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of organic acids, any acidic compound containing carbon in covalent linkage.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
aromatic compound catabolic process
The chemical reactions and pathways resulting in the breakdown of aromatic compounds, any substance containing an aromatic carbon ring.
drug catabolic process
The chemical reactions and pathways resulting in the breakdown of a drug, a substance used in the diagnosis, treatment or prevention of a disease.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
phospholipid metabolic process
The chemical reactions and pathways involving phospholipids, any lipid containing phosphoric acid as a mono- or diester.
cellular lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, as carried out by individual cells.
cellular carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, carried out by individual cells.
heterocycle catabolic process
The chemical reactions and pathways resulting in the breakdown of heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
polysaccharide metabolic process
The chemical reactions and pathways involving polysaccharides, a polymer of more than 20 monosaccharide residues joined by glycosidic linkages.
carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y.
cellular carbohydrate metabolic process
The chemical reactions and pathways involving carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, as carried out by individual cells.
lipid biosynthetic process
The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
monosaccharide metabolic process
The chemical reactions and pathways involving monosaccharides, the simplest carbohydrates. They are polyhydric alcohols containing either an aldehyde or a keto group and between three to ten or more carbon atoms. They form the constitutional repeating units of oligo- and polysaccharides.
alcohol biosynthetic process
The chemical reactions and pathways resulting in the formation of alcohols, any of a class of compounds containing one or more hydroxyl groups attached to a saturated carbon atom.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
activation of immune response
Any process that initiates an immune response.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
complement activation, alternative pathway
Any process involved in the activation of any of the steps of the alternative pathway of the complement cascade which allows for the direct killing of microbes and the regulation of other immune processes.
negative regulation of blood coagulation
Any process that stops, prevents or reduces the frequency, rate or extent of blood coagulation.
negative regulation of coagulation
Any process that stops, prevents or reduces the frequency, rate or extent of coagulation.
regulation of blood coagulation
Any process that modulates the frequency, rate or extent of blood coagulation.
positive regulation of coagulation
Any process that activates or increases the frequency, rate or extent of coagulation.
positive regulation of blood coagulation
Any process that activates or increases the frequency, rate or extent of blood coagulation.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells by means of some external agent such as a transporter or pore.
regulation of blood coagulation
Any process that modulates the frequency, rate or extent of blood coagulation.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
response to hormone stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hormone stimulus.
lipid transport
The directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore. Lipids are compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
regulation of lipid transport
Any process that modulates the frequency, rate or extent of the directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore.
negative regulation of lipid transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore.
hormone metabolic process
The chemical reactions and pathways involving any hormone, naturally occurring substances secreted by specialized cells that affects the metabolism or behavior of other cells possessing functional receptors for the hormone.
negative regulation of catalytic activity
Any process that stops or reduces the activity of an enzyme.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
amine biosynthetic process
The chemical reactions and pathways resulting in the formation of any organic compound that is weakly basic in character and contains an amino or a substituted amino group. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
exogenous drug catabolic process
The chemical reactions and pathways resulting in the breakdown of a drug that has originated externally to the cell or organism.
monosaccharide biosynthetic process
The chemical reactions and pathways resulting in the formation of monosaccharides, polyhydric alcohols containing either an aldehyde or a keto group and between three to ten or more carbon atoms.
negative regulation of hydrolase activity
Any process that stops or reduces the rate of hydrolase activity, the catalysis of the hydrolysis of various bonds.
sulfur amino acid metabolic process
The chemical reactions and pathways involving amino acids containing sulfur, comprising cysteine, homocysteine, methionine and selenocysteine.
cellular amino acid biosynthetic process
The chemical reactions and pathways resulting in the formation of amino acids, organic acids containing one or more amino substituents.
cellular amino acid catabolic process
The chemical reactions and pathways resulting in the breakdown of amino acids, organic acids containing one or more amino substituents.
cellular biogenic amine metabolic process
The chemical reactions and pathways occurring at the level of individual cells involving any of a group of naturally occurring, biologically active amines, such as norepinephrine, histamine, and serotonin, many of which act as neurotransmitters.
acylglycerol metabolic process
The chemical reactions and pathways involving acylglycerol, any mono-, di- or triester of glycerol with (one or more) fatty acids.
glycerophospholipid metabolic process
The chemical reactions and pathways involving glycerophospholipids, any derivative of glycerophosphate that contains at least one O-acyl, O-alkyl, or O-alkenyl group attached to the glycerol residue.
glycerolipid catabolic process
The chemical reactions and pathways resulting in the breakdown of glycerolipids, any lipid with a glycerol backbone.
monosaccharide biosynthetic process
The chemical reactions and pathways resulting in the formation of monosaccharides, polyhydric alcohols containing either an aldehyde or a keto group and between three to ten or more carbon atoms.
histidine catabolic process
The chemical reactions and pathways resulting in the breakdown of histidine, 2-amino-3-(1H-imidazol-4-yl)propanoic acid.
steroid biosynthetic process
The chemical reactions and pathways resulting in the formation of steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus; includes de novo formation and steroid interconversion by modification.
steroid catabolic process
The chemical reactions and pathways resulting in the breakdown of steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
sterol metabolic process
The chemical reactions and pathways involving sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
sterol biosynthetic process
The chemical reactions and pathways resulting in the formation of sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
phosphatidylcholine metabolic process
The chemical reactions and pathways involving phosphatidylcholines, any of a class of glycerophospholipids in which the phosphatidyl group is esterified to the hydroxyl group of choline. They are important constituents of cell membranes.
acylglycerol metabolic process
The chemical reactions and pathways involving acylglycerol, any mono-, di- or triester of glycerol with (one or more) fatty acids.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
complement activation, classical pathway
Any process involved in the activation of any of the steps of the classical pathway of the complement cascade which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes.
positive regulation of blood coagulation
Any process that activates or increases the frequency, rate or extent of blood coagulation.
negative regulation of blood coagulation
Any process that stops, prevents or reduces the frequency, rate or extent of blood coagulation.
positive regulation of fibrinolysis
Any process that activates, maintains or increases the frequency, rate or extent of fibrinolysis, an ongoing process that solubilizes fibrin, resulting in the removal of small blood clots.
negative regulation of lipid transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore.
regulation of blood coagulation
Any process that modulates the frequency, rate or extent of blood coagulation.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
regulation of wound healing
Any process that modulates the rate, frequency, or extent of the series of events that restore integrity to a damaged tissue, following an injury.
response to nutrient
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
regulation of sterol transport
Any process that modulates the frequency, rate or extent of the directed movement of sterols into, out of, within or between cells by means of some external agent such as a transporter or pore.
negative regulation of sterol transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of sterols into, out of, within or between cells by means of some external agent such as a transporter or pore.
ethanolamine and derivative metabolic process
The chemical reactions and pathways involving ethanolamine (2-aminoethanol) and compounds derived from it.
cellular amino acid catabolic process
The chemical reactions and pathways resulting in the breakdown of amino acids, organic acids containing one or more amino substituents.
bile acid catabolic process
The chemical reactions and pathways resulting in the breakdown of bile acids, any of a group of steroid carboxylic acids occurring in bile.
cellular amino acid biosynthetic process
The chemical reactions and pathways resulting in the formation of amino acids, organic acids containing one or more amino substituents.
negative regulation of lipase activity
Any process that decreases the frequency, rate or extent of lipase activity, the hydrolysis of a lipid or phospholipid.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
carboxylic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
cysteine metabolic process
The chemical reactions and pathways involving cysteine, 2-amino-3-mercaptopropanoic acid.
histidine metabolic process
The chemical reactions and pathways involving histidine, 2-amino-3-(1H-imidazol-4-yl)propanoic acid.
histidine family amino acid catabolic process
The chemical reactions and pathways resulting in the breakdown of amino acids of the histidine family.
hexose biosynthetic process
The chemical reactions and pathways resulting in the formation of hexose, any monosaccharide with a chain of six carbon atoms in the molecule.
bile acid catabolic process
The chemical reactions and pathways resulting in the breakdown of bile acids, any of a group of steroid carboxylic acids occurring in bile.
cholesterol biosynthetic process
The chemical reactions and pathways resulting in the formation of cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones.
regulation of fibrinolysis
Any process that modulates the frequency, rate or extent of fibrinolysis, an ongoing process that solubilizes fibrin, resulting in the removal of small blood clots.
positive regulation of fibrinolysis
Any process that activates, maintains or increases the frequency, rate or extent of fibrinolysis, an ongoing process that solubilizes fibrin, resulting in the removal of small blood clots.
negative regulation of sterol transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of sterols into, out of, within or between cells by means of some external agent such as a transporter or pore.
regulation of cholesterol transport
Any process that modulates the frequency, rate or extent of the directed movement of cholesterol into, out of, within or between cells by means of some external agent such as a transporter or pore.
negative regulation of cholesterol transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of cholesterol into, out of, within or between cells by means of some external agent such as a transporter or pore.
histidine catabolic process
The chemical reactions and pathways resulting in the breakdown of histidine, 2-amino-3-(1H-imidazol-4-yl)propanoic acid.
negative regulation of lipoprotein lipase activity
Any process that stops or reduces the activity of the enzyme lipoprotein lipase.
activation of plasma proteins involved in acute inflammatory response
Any process activating plasma proteins by proteolysis as part of an acute inflammatory response.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
glyoxylate metabolic process
The chemical reactions and pathways involving glyoxylate, the anion of glyoxylic acid, HOC-COOH.
gluconeogenesis
The formation of glucose from noncarbohydrate precursors, such as pyruvate, amino acids and glycerol.
humoral immune response mediated by circulating immunoglobulin
An immune response dependent upon secreted immunoglobulin. An example of this process is found in Mus musculus.
negative regulation of cholesterol transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of cholesterol into, out of, within or between cells by means of some external agent such as a transporter or pore.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
gluconeogenesis
The formation of glucose from noncarbohydrate precursors, such as pyruvate, amino acids and glycerol.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell fraction
A generic term for parts of cells prepared by disruptive biochemical techniques.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
fibrinogen complex
A highly soluble, elongated protein complex found in blood plasma and involved in clot formation. It is converted into fibrin monomer by the action of thrombin. In the mouse, fibrinogen is a hexamer, 46 nm long and 9 nm maximal diameter, containing two sets of nonidentical chains (alpha, beta, and gamma) linked together by disulfide bonds.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
membrane fraction
That fraction of cells, prepared by disruptive biochemical methods, that includes the plasma and other membranes.
insoluble fraction
That fraction of cells, prepared by disruptive biochemical methods, that is not soluble in water.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
microsome
Any of the small, heterogeneous, artifactual, vesicular particles, 50-150 nm in diameter, that are formed when some eukaryotic cells are homogenized and that sediment on centrifugation at 100000 g.
stored secretory granule
A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell and upon stimulation, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
organelle membrane
The lipid bilayer surrounding an organelle.
platelet alpha granule
A secretory organelle found in blood platelets, which is unique in that it exhibits further compartmentalization and acquires its protein content via two distinct mechanisms: (1) biosynthesis predominantly at the megakaryocyte (MK) level (with some vestigial platelet synthesis) (e.g. platelet factor 4) and (2) endocytosis and pinocytosis at both the MK and circulating platelet levels (e.g. fibrinogen (Fg) and IgG).
platelet alpha granule lumen
The volume enclosed by the membrane of the platelet alpha granule.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
vesicle lumen
The volume enclosed by the membrane or protein that forms a vesicle.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
protein-lipid complex
A macromolecular complex containing both protein and lipid molecules.
plasma lipoprotein particle
A spherical particle with a hydrophobic core of triglycerides and/or cholesterol esters, surrounded by an amphipathic monolayer of phospholipids, cholesterol and apolipoproteins. Plasma lipoprotein particles transport lipids, which are non-covalently associated with the particles, in the blood or lymph.
high-density lipoprotein particle
A lipoprotein particle with a high density (typically 1.063-1.21 g/ml) and a diameter of 5-10 nm that contains APOAs and may contain APOCs and APOE; found in blood and carries lipids from body tissues to the liver as part of the reverse cholesterol transport process.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
vesicular fraction
Any of the small, heterogeneous, artifactual, vesicular particles that are formed when some cells are homogenized.
chylomicron
A large lipoprotein particle (diameter 75-1200 nm) composed of a central core of triglycerides and cholesterol surrounded by a protein-phospholipid coating. The proteins include one molecule of apolipoprotein B-48 and may include a variety of apolipoproteins, including APOAs, APOCs and APOE. Chylomicrons are found in blood or lymph and carry lipids from the intestines into other body tissues.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic membrane-bounded vesicle lumen
The volume enclosed by the membrane of a cytoplasmic membrane-bounded vesicle.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
plasma lipoprotein particle
A spherical particle with a hydrophobic core of triglycerides and/or cholesterol esters, surrounded by an amphipathic monolayer of phospholipids, cholesterol and apolipoproteins. Plasma lipoprotein particles transport lipids, which are non-covalently associated with the particles, in the blood or lymph.
fibrinogen complex
A highly soluble, elongated protein complex found in blood plasma and involved in clot formation. It is converted into fibrin monomer by the action of thrombin. In the mouse, fibrinogen is a hexamer, 46 nm long and 9 nm maximal diameter, containing two sets of nonidentical chains (alpha, beta, and gamma) linked together by disulfide bonds.
organelle membrane
The lipid bilayer surrounding an organelle.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
fibrinogen complex
A highly soluble, elongated protein complex found in blood plasma and involved in clot formation. It is converted into fibrin monomer by the action of thrombin. In the mouse, fibrinogen is a hexamer, 46 nm long and 9 nm maximal diameter, containing two sets of nonidentical chains (alpha, beta, and gamma) linked together by disulfide bonds.
plasma lipoprotein particle
A spherical particle with a hydrophobic core of triglycerides and/or cholesterol esters, surrounded by an amphipathic monolayer of phospholipids, cholesterol and apolipoproteins. Plasma lipoprotein particles transport lipids, which are non-covalently associated with the particles, in the blood or lymph.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
vesicle lumen
The volume enclosed by the membrane or protein that forms a vesicle.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
cytoplasmic membrane-bounded vesicle lumen
The volume enclosed by the membrane of a cytoplasmic membrane-bounded vesicle.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoplasmic membrane-bounded vesicle lumen
The volume enclosed by the membrane of a cytoplasmic membrane-bounded vesicle.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
platelet alpha granule lumen
The volume enclosed by the membrane of the platelet alpha granule.

electron carrier activity
Any molecular entity that serves as an electron acceptor and electron donor in an electron transport system.
protein binding
Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
pattern binding
Interacting selectively and non-covalently with a repeating or polymeric structure, such as a polysaccharide or peptidoglycan.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
serine-type endopeptidase activity
Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
endopeptidase activity
Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain.
serine-type peptidase activity
Catalysis of the hydrolysis of peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
peptidase activity
Catalysis of the hydrolysis of a peptide bond. A peptide bond is a covalent bond formed when the carbon atom from the carboxyl group of one amino acid shares electrons with the nitrogen atom from the amino group of a second amino acid.
monooxygenase activity
Catalysis of the incorporation of one atom from molecular oxygen into a compound and the reduction of the other atom of oxygen to water.
enzyme inhibitor activity
Stops, prevents or reduces the activity of an enzyme.
endopeptidase inhibitor activity
Stops, prevents or reduces the activity of an endopeptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
serine-type endopeptidase inhibitor activity
Stops, prevents or reduces the activity of serine-type endopeptidases, enzymes that catalyze the hydrolysis of nonterminal peptide bonds in a polypeptide chain; a serine residue (and a histidine residue) are at the active center of the enzyme.
carbohydrate binding
Interacting selectively and non-covalently with any carbohydrate, any of a group of organic compounds based of the general formula Cx(H2O)y.
growth factor binding
Interacting selectively and non-covalently with any growth factor, proteins or polypeptides that stimulate a cell or organism to grow or proliferate.
neurotrophin binding
Interacting selectively and non-covalently with neurotrophin, any of a family of growth factors that block apoptosis in neurons and thus promote nerve growth.
nerve growth factor binding
Interacting selectively and non-covalently with nerve growth factor.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
organic acid transmembrane transporter activity
Catalysis of the transfer of organic acids, any acidic compound containing carbon in covalent linkage, from one side of the membrane to the other.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
steroid binding
Interacting selectively and non-covalently with a steroid, any of a large group of substances that have in common a ring system based on 1,2-cyclopentanoperhydrophenanthrene.
metal ion binding
Interacting selectively and non-covalently with any metal ion.
iron ion binding
Interacting selectively and non-covalently with iron (Fe) ions.
glycosaminoglycan binding
Interacting selectively and non-covalently with any glycan (polysaccharide) containing a substantial proportion of aminomonosaccharide residues.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
lipid binding
Interacting selectively and non-covalently with a lipid.
anion transmembrane transporter activity
Catalysis of the transfer of a negatively charged ion from one side of a membrane to the other.
organic anion transmembrane transporter activity
Catalysis of the transfer of organic anions from one side of a membrane to the other. Organic anions are atoms or small molecules with a negative charge which contain carbon in covalent linkage.
peptidase activity, acting on L-amino acid peptides
Catalysis of the hydrolysis of peptide bonds formed between L-amino acids.
endopeptidase regulator activity
Modulates the activity of a peptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
oxygen binding
Interacting selectively and non-covalently with oxygen (O2).
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
amino acid binding
Interacting selectively and non-covalently with an amino acid, organic acids containing one or more amino substituents.
oxidoreductase activity, acting on the CH-CH group of donors
Catalysis of an oxidation-reduction (redox) reaction in which a CH-CH group acts as a hydrogen or electron donor and reduces a hydrogen or electron acceptor.
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen
Catalysis of an oxidation-reduction (redox) reaction in which hydrogen or electrons are transferred from each of two donors, and molecular oxygen is reduced or incorporated into a donor.
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen
Catalysis of an oxidation-reduction (redox) reaction in which hydrogen or electrons are transferred from reduced flavin or flavoprotein and one other donor, and one atom of oxygen is incorporated into one donor.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
lyase activity
Catalysis of the cleavage of C-C, C-O, C-N and other bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond. They differ from other enzymes in that two substrates are involved in one reaction direction, but only one in the other direction. When acting on the single substrate, a molecule is eliminated and this generates either a new double bond or a new ring.
carbon-nitrogen lyase activity
Catalysis of the release of ammonia or one of its derivatives, with the formation of a double bond or ring. Enzymes with this activity may catalyze the actual elimination of the ammonia, amine or amide, e.g. CH-CH(-NH-R) = C=CH- + NH2-R. Others, however, catalyze elimination of another component, e.g. water, which is followed by spontaneous reactions that lead to breakage of the C-N bond, e.g. L-serine ammonia-lyase (EC:4.3.1.17), so that the overall reaction is C(-OH)-CH(-NH2) = CH2-CO- + NH3, i.e. an elimination with rearrangement. The sub-subclasses of EC:4.3 are the ammonia-lyases (EC:4.3.1), lyases acting on amides, amidines, etc. (EC:4.3.2), the amine-lyases (EC:4.3.3), and other carbon-nitrogen lyases (EC:4.3.99).
ammonia-lyase activity
Catalysis of the release of ammonia by the cleavage of a carbon-nitrogen bond or the reverse reaction with ammonia as a substrate.
peptidase inhibitor activity
Stops, prevents or reduces the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
serine hydrolase activity
Catalysis of the hydrolysis of a substrate by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
vitamin binding
Interacting selectively and non-covalently with a vitamin, one of a number of unrelated organic substances that occur in many foods in small amounts and that are necessary in trace amounts for the normal metabolic functioning of the body.
heme binding
Interacting selectively and non-covalently with heme, any compound of iron complexed in a porphyrin (tetrapyrrole) ring.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
pyridoxal phosphate binding
Interacting selectively and non-covalently with pyridoxal 5' phosphate, 3-hydroxy-5-(hydroxymethyl)-2-methyl4-pyridine carboxaldehyde 5' phosphate, the biologically active form of vitamin B6.
enzyme regulator activity
Modulates the activity of an enzyme.
polysaccharide binding
Interacting selectively and non-covalently with any polysaccharide, a polymer of more than 20 monosaccharide residues joined by glycosidic linkages.
carboxylic acid binding
Interacting selectively and non-covalently with a carboxylic acid, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
ion binding
Interacting selectively and non-covalently with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively and non-covalently with cations, charged atoms or groups of atoms with a net positive charge.
amine binding
Interacting selectively and non-covalently with any organic compound that is weakly basic in character and contains an amino or a substituted amino group.
tetrapyrrole binding
Interacting selectively and non-covalently with a tetrapyrrole, a compound containing four pyrrole nuclei variously substituted and linked to each other through carbons at the alpha position.
transition metal ion binding
Interacting selectively and non-covalently with a transition metal ions; a transition metal is an element whose atom has an incomplete d-subshell of extranuclear electrons, or which gives rise to a cation or cations with an incomplete d-subshell. Transition metals often have more than one valency state. Biologically relevant transition metals include vanadium, manganese, iron, copper, cobalt, nickel, molybdenum and silver.
carboxylic acid transmembrane transporter activity
Catalysis of the transfer of carboxylic acids from one side of the membrane to the other. Carboxylic acids are organic acids containing one or more carboxyl (COOH) groups or anions (COO-).
cofactor binding
Interacting selectively and non-covalently with a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
coenzyme binding
Interacting selectively and non-covalently with a coenzyme, any of various nonprotein organic cofactors that are required, in addition to an enzyme and a substrate, for an enzymatic reaction to proceed.
peptidase regulator activity
Modulates the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
vitamin B6 binding
Interacting selectively and non-covalently with any of the vitamin B6 compounds: pyridoxal, pyridoxamine and pyridoxine and the active form, pyridoxal phosphate.
aromatase activity
Catalysis of the reduction of an aliphatic ring to yield an aromatic ring.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
polysaccharide binding
Interacting selectively and non-covalently with any polysaccharide, a polymer of more than 20 monosaccharide residues joined by glycosidic linkages.
amino acid binding
Interacting selectively and non-covalently with an amino acid, organic acids containing one or more amino substituents.
peptidase inhibitor activity
Stops, prevents or reduces the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen
Catalysis of an oxidation-reduction (redox) reaction in which hydrogen or electrons are transferred from reduced flavin or flavoprotein and one other donor, and one atom of oxygen is incorporated into one donor.
pyridoxal phosphate binding
Interacting selectively and non-covalently with pyridoxal 5' phosphate, 3-hydroxy-5-(hydroxymethyl)-2-methyl4-pyridine carboxaldehyde 5' phosphate, the biologically active form of vitamin B6.
endopeptidase inhibitor activity
Stops, prevents or reduces the activity of an endopeptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
serine-type peptidase activity
Catalysis of the hydrolysis of peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
serine-type endopeptidase activity
Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
heme binding
Interacting selectively and non-covalently with heme, any compound of iron complexed in a porphyrin (tetrapyrrole) ring.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04610 | 2.108e-16 | 2.033 | 23 | 50 | Complement and coagulation cascades |
| 01100 | 6.486e-08 | 32.74 | 66 | 805 | Metabolic pathways |
| 00830 | 1.420e-06 | 1.017 | 10 | 25 | Retinol metabolism |
| 00260 | 1.266e-04 | 0.976 | 8 | 24 | Glycine, serine and threonine metabolism |
| 04950 | 6.016e-04 | 0.61 | 6 | 15 | Maturity onset diabetes of the young |
| 00980 | 1.820e-03 | 1.057 | 7 | 26 | Metabolism of xenobiotics by cytochrome P450 |
| 00982 | 2.292e-03 | 1.098 | 7 | 27 | Drug metabolism - cytochrome P450 |
| 00140 | 4.970e-03 | 0.8947 | 6 | 22 | Steroid hormone biosynthesis |
| 00232 | 6.648e-03 | 0.1627 | 3 | 4 | Caffeine metabolism |
| 00340 | 7.588e-03 | 0.976 | 6 | 24 | Histidine metabolism |
| 05020 | 7.588e-03 | 0.976 | 6 | 24 | Prion diseases |
| 00120 | 1.055e-02 | 0.4067 | 4 | 10 | Primary bile acid biosynthesis |
| 00500 | 2.469e-02 | 0.854 | 5 | 21 | Starch and sucrose metabolism |
| 00380 | 2.904e-02 | 1.301 | 6 | 32 | Tryptophan metabolism |
| 00270 | 3.452e-02 | 0.9353 | 5 | 23 | Cysteine and methionine metabolism |
| 00430 | 3.601e-02 | 0.2847 | 3 | 7 | Taurine and hypotaurine metabolism |
A1CFAPOBEC1 complementation factor (ENSG00000148584), score: 0.82 ABCB11ATP-binding cassette, sub-family B (MDR/TAP), member 11 (ENSG00000073734), score: 0.75 ABCC2ATP-binding cassette, sub-family C (CFTR/MRP), member 2 (ENSG00000023839), score: 0.77 ABCC3ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (ENSG00000108846), score: 0.72 ABCG5ATP-binding cassette, sub-family G (WHITE), member 5 (ENSG00000138075), score: 0.92 ACOT12acyl-CoA thioesterase 12 (ENSG00000172497), score: 0.87 ACOX1acyl-CoA oxidase 1, palmitoyl (ENSG00000161533), score: 0.83 ACP2acid phosphatase 2, lysosomal (ENSG00000134575), score: 0.77 ACSM5acyl-CoA synthetase medium-chain family member 5 (ENSG00000183549), score: 0.78 ADH4alcohol dehydrogenase 4 (class II), pi polypeptide (ENSG00000198099), score: 0.91 ADH6alcohol dehydrogenase 6 (class V) (ENSG00000172955), score: 0.78 ADKadenosine kinase (ENSG00000156110), score: 0.82 ADRA1Aadrenergic, alpha-1A-, receptor (ENSG00000120907), score: 0.74 AFMafamin (ENSG00000079557), score: 0.82 AGFG2ArfGAP with FG repeats 2 (ENSG00000106351), score: 0.82 AGTR1angiotensin II receptor, type 1 (ENSG00000144891), score: 0.75 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.88 AHRaryl hydrocarbon receptor (ENSG00000106546), score: 0.77 AIG1androgen-induced 1 (ENSG00000146416), score: 0.75 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.94 ALAS1aminolevulinate, delta-, synthase 1 (ENSG00000023330), score: 0.83 ALBalbumin (ENSG00000163631), score: 0.83 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (ENSG00000165092), score: 0.74 ALDH1B1aldehyde dehydrogenase 1 family, member B1 (ENSG00000137124), score: 0.75 ALDH2aldehyde dehydrogenase 2 family (mitochondrial) (ENSG00000111275), score: 0.74 AMBPalpha-1-microglobulin/bikunin precursor (ENSG00000106927), score: 0.86 AMDHD1amidohydrolase domain containing 1 (ENSG00000139344), score: 0.85 ANK2ankyrin 2, neuronal (ENSG00000145362), score: -0.74 ANO1anoctamin 1, calcium activated chloride channel (ENSG00000131620), score: 0.76 APCSamyloid P component, serum (ENSG00000132703), score: 0.84 APOBapolipoprotein B (including Ag(x) antigen) (ENSG00000084674), score: 0.88 APOC1apolipoprotein C-I (ENSG00000130208), score: 0.88 APOC3apolipoprotein C-III (ENSG00000110245), score: 0.87 APOFapolipoprotein F (ENSG00000175336), score: 0.87 AQP9aquaporin 9 (ENSG00000103569), score: 0.9 ARG1arginase, liver (ENSG00000118520), score: 0.96 ARHGEF26Rho guanine nucleotide exchange factor (GEF) 26 (ENSG00000114790), score: 0.77 ART4ADP-ribosyltransferase 4 (Dombrock blood group) (ENSG00000111339), score: 0.89 ASPGasparaginase homolog (S. cerevisiae) (ENSG00000166183), score: 0.78 ATP11CATPase, class VI, type 11C (ENSG00000101974), score: 0.74 AUP1ancient ubiquitous protein 1 (ENSG00000115307), score: 0.75 BAATbile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) (ENSG00000136881), score: 0.86 BCHEbutyrylcholinesterase (ENSG00000114200), score: 0.81 BOCBoc homolog (mouse) (ENSG00000144857), score: -0.74 BTDbiotinidase (ENSG00000169814), score: 0.72 C10orf47chromosome 10 open reading frame 47 (ENSG00000148426), score: 0.75 C14orf21chromosome 14 open reading frame 21 (ENSG00000196943), score: 0.88 C15orf59chromosome 15 open reading frame 59 (ENSG00000205363), score: -0.77 C1orf130chromosome 1 open reading frame 130 (ENSG00000184454), score: 0.79 C20orf194chromosome 20 open reading frame 194 (ENSG00000088854), score: -0.82 C4BPAcomplement component 4 binding protein, alpha (ENSG00000123838), score: 0.87 C4BPBcomplement component 4 binding protein, beta (ENSG00000123843), score: 0.86 C4orf19chromosome 4 open reading frame 19 (ENSG00000154274), score: 0.72 C4orf34chromosome 4 open reading frame 34 (ENSG00000163683), score: 0.76 C5complement component 5 (ENSG00000106804), score: 0.85 C5orf33chromosome 5 open reading frame 33 (ENSG00000152620), score: 0.88 C6complement component 6 (ENSG00000039537), score: 0.74 C8Acomplement component 8, alpha polypeptide (ENSG00000157131), score: 0.91 C8Bcomplement component 8, beta polypeptide (ENSG00000021852), score: 0.89 C8Gcomplement component 8, gamma polypeptide (ENSG00000176919), score: 0.81 C9complement component 9 (ENSG00000113600), score: 0.88 CATcatalase (ENSG00000121691), score: 0.74 CCDC152coiled-coil domain containing 152 (ENSG00000198865), score: 0.78 CD14CD14 molecule (ENSG00000170458), score: 0.74 CD247CD247 molecule (ENSG00000198821), score: 0.72 CD4CD4 molecule (ENSG00000010610), score: 0.78 CDO1cysteine dioxygenase, type I (ENSG00000129596), score: 0.74 CEACAM16carcinoembryonic antigen-related cell adhesion molecule 16 (ENSG00000213892), score: 0.81 CETPcholesteryl ester transfer protein, plasma (ENSG00000087237), score: 0.72 CFHcomplement factor H (ENSG00000000971), score: 0.73 CFPcomplement factor properdin (ENSG00000126759), score: 0.84 CHST10carbohydrate sulfotransferase 10 (ENSG00000115526), score: -0.78 CHST13carbohydrate (chondroitin 4) sulfotransferase 13 (ENSG00000180767), score: 0.79 CIDEBcell death-inducing DFFA-like effector b (ENSG00000136305), score: 0.88 CLDN1claudin 1 (ENSG00000163347), score: 0.78 CLDN14claudin 14 (ENSG00000159261), score: 0.89 CLEC1BC-type lectin domain family 1, member B (ENSG00000165682), score: 0.86 CMTM8CKLF-like MARVEL transmembrane domain containing 8 (ENSG00000170293), score: 0.74 CNPY3canopy 3 homolog (zebrafish) (ENSG00000137161), score: 0.74 COLEC10collectin sub-family member 10 (C-type lectin) (ENSG00000184374), score: 0.79 CPceruloplasmin (ferroxidase) (ENSG00000047457), score: 0.79 CPB2carboxypeptidase B2 (plasma) (ENSG00000080618), score: 0.89 CPN2carboxypeptidase N, polypeptide 2 (ENSG00000178772), score: 0.8 CREB3L3cAMP responsive element binding protein 3-like 3 (ENSG00000060566), score: 0.87 CREG1cellular repressor of E1A-stimulated genes 1 (ENSG00000143162), score: 0.72 CSADcysteine sulfinic acid decarboxylase (ENSG00000139631), score: 0.73 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.73 CXCR2chemokine (C-X-C motif) receptor 2 (ENSG00000180871), score: 0.76 CYP17A1cytochrome P450, family 17, subfamily A, polypeptide 1 (ENSG00000148795), score: 0.73 CYP1A1cytochrome P450, family 1, subfamily A, polypeptide 1 (ENSG00000140465), score: 0.76 CYP1A2cytochrome P450, family 1, subfamily A, polypeptide 2 (ENSG00000140505), score: 0.95 CYP2C8cytochrome P450, family 2, subfamily C, polypeptide 8 (ENSG00000138115), score: 0.86 CYP39A1cytochrome P450, family 39, subfamily A, polypeptide 1 (ENSG00000146233), score: 0.84 CYP3A43cytochrome P450, family 3, subfamily A, polypeptide 43 (ENSG00000021461), score: 0.98 CYP4F11cytochrome P450, family 4, subfamily F, polypeptide 11 (ENSG00000171903), score: 0.74 DBN1drebrin 1 (ENSG00000113758), score: -0.76 DDI2DNA-damage inducible 1 homolog 2 (S. cerevisiae) (ENSG00000197312), score: 0.73 DGAT2diacylglycerol O-acyltransferase 2 (ENSG00000062282), score: 0.78 DHCR77-dehydrocholesterol reductase (ENSG00000172893), score: 0.72 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.83 DMRTA1DMRT-like family A1 (ENSG00000176399), score: 0.75 DNAJA4DnaJ (Hsp40) homolog, subfamily A, member 4 (ENSG00000140403), score: -0.74 DNAJC22DnaJ (Hsp40) homolog, subfamily C, member 22 (ENSG00000178401), score: 0.72 DNASE1L3deoxyribonuclease I-like 3 (ENSG00000163687), score: 0.79 DPYDdihydropyrimidine dehydrogenase (ENSG00000188641), score: 0.85 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (ENSG00000134109), score: 0.75 EI24etoposide induced 2.4 mRNA (ENSG00000149547), score: 0.9 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENSG00000197594), score: 0.75 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (ENSG00000143819), score: 0.87 ERLIN1ER lipid raft associated 1 (ENSG00000107566), score: 0.74 ERP29endoplasmic reticulum protein 29 (ENSG00000089248), score: 0.74 ESR1estrogen receptor 1 (ENSG00000091831), score: 0.88 F10coagulation factor X (ENSG00000126218), score: 0.8 F11coagulation factor XI (ENSG00000088926), score: 0.84 F12coagulation factor XII (Hageman factor) (ENSG00000131187), score: 0.77 F2coagulation factor II (thrombin) (ENSG00000180210), score: 0.88 F7coagulation factor VII (serum prothrombin conversion accelerator) (ENSG00000057593), score: 0.9 F9coagulation factor IX (ENSG00000101981), score: 0.89 FCN2ficolin (collagen/fibrinogen domain containing lectin) 2 (hucolin) (ENSG00000160339), score: 0.75 FETUBfetuin B (ENSG00000090512), score: 0.97 FGBfibrinogen beta chain (ENSG00000171564), score: 0.79 FGGfibrinogen gamma chain (ENSG00000171557), score: 0.8 FMO3flavin containing monooxygenase 3 (ENSG00000007933), score: 0.87 FMO5flavin containing monooxygenase 5 (ENSG00000131781), score: 0.84 FN1fibronectin 1 (ENSG00000115414), score: 0.73 FOXA1forkhead box A1 (ENSG00000129514), score: 0.9 FOXA3forkhead box A3 (ENSG00000170608), score: 0.89 FRRS1ferric-chelate reductase 1 (ENSG00000156869), score: 0.75 FSTfollistatin (ENSG00000134363), score: 0.74 FTCDformiminotransferase cyclodeaminase (ENSG00000160282), score: 0.76 FURINfurin (paired basic amino acid cleaving enzyme) (ENSG00000140564), score: 0.8 GALTgalactose-1-phosphate uridylyltransferase (ENSG00000213930), score: 0.78 GAMTguanidinoacetate N-methyltransferase (ENSG00000130005), score: 0.74 GCgroup-specific component (vitamin D binding protein) (ENSG00000145321), score: 0.85 GCKglucokinase (hexokinase 4) (ENSG00000106633), score: 0.9 GCKRglucokinase (hexokinase 4) regulator (ENSG00000084734), score: 0.87 GCLCglutamate-cysteine ligase, catalytic subunit (ENSG00000001084), score: 0.83 GDF2growth differentiation factor 2 (ENSG00000128802), score: 0.88 GGCXgamma-glutamyl carboxylase (ENSG00000115486), score: 0.79 GHRgrowth hormone receptor (ENSG00000112964), score: 0.82 GLYATL3glycine-N-acyltransferase-like 3 (ENSG00000203972), score: 0.87 GNEglucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase (ENSG00000159921), score: 0.88 GNMTglycine N-methyltransferase (ENSG00000124713), score: 0.93 GNPNAT1glucosamine-phosphate N-acetyltransferase 1 (ENSG00000100522), score: 0.82 GPAMglycerol-3-phosphate acyltransferase, mitochondrial (ENSG00000119927), score: 0.91 GPLD1glycosylphosphatidylinositol specific phospholipase D1 (ENSG00000112293), score: 0.73 GPR128G protein-coupled receptor 128 (ENSG00000144820), score: 0.97 GPTglutamic-pyruvate transaminase (alanine aminotransferase) (ENSG00000167701), score: 0.79 GUCA2Bguanylate cyclase activator 2B (uroguanylin) (ENSG00000044012), score: 0.91 GYS2glycogen synthase 2 (liver) (ENSG00000111713), score: 0.97 HALhistidine ammonia-lyase (ENSG00000084110), score: 0.8 HAO1hydroxyacid oxidase (glycolate oxidase) 1 (ENSG00000101323), score: 0.93 HDGFRP3hepatoma-derived growth factor, related protein 3 (ENSG00000166503), score: -0.76 HEBP1heme binding protein 1 (ENSG00000013583), score: 0.74 HMGCS23-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) (ENSG00000134240), score: 0.83 HPXhemopexin (ENSG00000110169), score: 0.86 HRGhistidine-rich glycoprotein (ENSG00000113905), score: 0.85 HS3ST3B1heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 (ENSG00000125430), score: 0.77 HSD11B1hydroxysteroid (11-beta) dehydrogenase 1 (ENSG00000117594), score: 0.83 HSD17B13hydroxysteroid (17-beta) dehydrogenase 13 (ENSG00000170509), score: 0.86 IDH1isocitrate dehydrogenase 1 (NADP+), soluble (ENSG00000138413), score: 0.72 IFT20intraflagellar transport 20 homolog (Chlamydomonas) (ENSG00000109083), score: 0.77 IGF1insulin-like growth factor 1 (somatomedin C) (ENSG00000017427), score: 0.77 IGF1Rinsulin-like growth factor 1 receptor (ENSG00000140443), score: -0.81 IGFALSinsulin-like growth factor binding protein, acid labile subunit (ENSG00000099769), score: 0.92 IGFBP1insulin-like growth factor binding protein 1 (ENSG00000146678), score: 0.8 IL1RAPinterleukin 1 receptor accessory protein (ENSG00000196083), score: 0.71 IL22RA1interleukin 22 receptor, alpha 1 (ENSG00000142677), score: 0.76 IL2RBinterleukin 2 receptor, beta (ENSG00000100385), score: 0.81 IL6Rinterleukin 6 receptor (ENSG00000160712), score: 0.81 INHBCinhibin, beta C (ENSG00000175189), score: 0.91 INHBEinhibin, beta E (ENSG00000139269), score: 0.89 INSIG1insulin induced gene 1 (ENSG00000186480), score: 0.83 ITIH1inter-alpha (globulin) inhibitor H1 (ENSG00000055957), score: 0.88 ITIH3inter-alpha (globulin) inhibitor H3 (ENSG00000162267), score: 0.8 ITIH4inter-alpha (globulin) inhibitor H4 (plasma Kallikrein-sensitive glycoprotein) (ENSG00000055955), score: 0.82 IVDisovaleryl-CoA dehydrogenase (ENSG00000128928), score: 0.72 KDM3Blysine (K)-specific demethylase 3B (ENSG00000120733), score: -0.75 KLBklotho beta (ENSG00000134962), score: 0.73 KLKB1kallikrein B, plasma (Fletcher factor) 1 (ENSG00000164344), score: 0.88 LASS2LAG1 homolog, ceramide synthase 2 (ENSG00000143418), score: 0.72 LBPlipopolysaccharide binding protein (ENSG00000129988), score: 0.78 LEAP2liver expressed antimicrobial peptide 2 (ENSG00000164406), score: 0.72 LECT2leukocyte cell-derived chemotaxin 2 (ENSG00000145826), score: 0.96 LEPRleptin receptor (ENSG00000116678), score: 0.78 LIPClipase, hepatic (ENSG00000166035), score: 0.89 LOC55908hepatocellular carcinoma-associated gene TD26 (ENSG00000130173), score: 0.83 LRG1leucine-rich alpha-2-glycoprotein 1 (ENSG00000171236), score: 0.8 LRRC3leucine rich repeat containing 3 (ENSG00000160233), score: 0.81 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.74 MAT1Amethionine adenosyltransferase I, alpha (ENSG00000151224), score: 0.9 MBNL3muscleblind-like 3 (Drosophila) (ENSG00000076770), score: 0.73 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.82 MOCOSmolybdenum cofactor sulfurase (ENSG00000075643), score: 0.79 MTHFD1methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase (ENSG00000100714), score: 0.79 MTTPmicrosomal triglyceride transfer protein (ENSG00000138823), score: 0.8 MUTmethylmalonyl CoA mutase (ENSG00000146085), score: 0.85 MYEF2myelin expression factor 2 (ENSG00000104177), score: -0.74 NAAAN-acylethanolamine acid amidase (ENSG00000138744), score: 0.77 NADKNAD kinase (ENSG00000008130), score: 0.74 NAT2N-acetyltransferase 2 (arylamine N-acetyltransferase) (ENSG00000156006), score: 0.91 NIPAL1NIPA-like domain containing 1 (ENSG00000163293), score: 0.84 NPC1L1NPC1 (Niemann-Pick disease, type C1, gene)-like 1 (ENSG00000015520), score: 0.96 NR1I2nuclear receptor subfamily 1, group I, member 2 (ENSG00000144852), score: 0.94 NR1I3nuclear receptor subfamily 1, group I, member 3 (ENSG00000143257), score: 0.79 NR5A2nuclear receptor subfamily 5, group A, member 2 (ENSG00000116833), score: 0.85 NT5M5',3'-nucleotidase, mitochondrial (ENSG00000205309), score: -0.75 OIT3oncoprotein induced transcript 3 (ENSG00000138315), score: 0.79 ONECUT1one cut homeobox 1 (ENSG00000169856), score: 0.83 ONECUT2one cut homeobox 2 (ENSG00000119547), score: 0.8 ORM1orosomucoid 1 (ENSG00000229314), score: 0.79 ORMDL3ORM1-like 3 (S. cerevisiae) (ENSG00000172057), score: 0.74 OTCornithine carbamoyltransferase (ENSG00000036473), score: 1 OXCT13-oxoacid CoA transferase 1 (ENSG00000083720), score: -0.74 OXER1oxoeicosanoid (OXE) receptor 1 (ENSG00000162881), score: 0.75 P4HBprolyl 4-hydroxylase, beta polypeptide (ENSG00000185624), score: 0.75 PACS1phosphofurin acidic cluster sorting protein 1 (ENSG00000175115), score: -0.75 PAHphenylalanine hydroxylase (ENSG00000171759), score: 0.74 PCSK6proprotein convertase subtilisin/kexin type 6 (ENSG00000140479), score: 0.78 PEMTphosphatidylethanolamine N-methyltransferase (ENSG00000133027), score: 0.9 PFKFB16-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (ENSG00000158571), score: 0.9 PGLYRP2peptidoglycan recognition protein 2 (ENSG00000161031), score: 0.95 PKLRpyruvate kinase, liver and RBC (ENSG00000143627), score: 0.74 PLA2G12Bphospholipase A2, group XIIB (ENSG00000138308), score: 0.75 PON1paraoxonase 1 (ENSG00000005421), score: 0.92 PON3paraoxonase 3 (ENSG00000105852), score: 0.85 PORP450 (cytochrome) oxidoreductase (ENSG00000127948), score: 0.82 PRG4proteoglycan 4 (ENSG00000116690), score: 0.76 PROCprotein C (inactivator of coagulation factors Va and VIIIa) (ENSG00000115718), score: 0.81 PROZprotein Z, vitamin K-dependent plasma glycoprotein (ENSG00000126231), score: 0.75 PRRG4proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) (ENSG00000135378), score: 0.76 PTPLBprotein tyrosine phosphatase-like (proline instead of catalytic arginine), member b (ENSG00000206527), score: 0.73 PYGLphosphorylase, glycogen, liver (ENSG00000100504), score: 0.84 RAPH1Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 (ENSG00000173166), score: 0.86 RARRES2retinoic acid receptor responder (tazarotene induced) 2 (ENSG00000106538), score: 0.81 RBP4retinol binding protein 4, plasma (ENSG00000138207), score: 0.73 RCL1RNA terminal phosphate cyclase-like 1 (ENSG00000120158), score: 0.76 RDH16retinol dehydrogenase 16 (all-trans) (ENSG00000139547), score: 0.91 RDH5retinol dehydrogenase 5 (11-cis/9-cis) (ENSG00000135437), score: 0.8 RNF130ring finger protein 130 (ENSG00000113269), score: 0.76 RNF185ring finger protein 185 (ENSG00000138942), score: 0.73 RTP3receptor (chemosensory) transporter protein 3 (ENSG00000163825), score: 0.78 SARDHsarcosine dehydrogenase (ENSG00000123453), score: 0.77 SCARB1scavenger receptor class B, member 1 (ENSG00000073060), score: 0.77 SCP2sterol carrier protein 2 (ENSG00000116171), score: 0.74 SDC2syndecan 2 (ENSG00000169439), score: 0.81 SDSserine dehydratase (ENSG00000135094), score: 0.8 SEBOXSEBOX homeobox (ENSG00000109072), score: 0.79 SEC14L2SEC14-like 2 (S. cerevisiae) (ENSG00000100003), score: 0.73 SEC14L3SEC14-like 3 (S. cerevisiae) (ENSG00000100012), score: 0.94 SEC14L4SEC14-like 4 (S. cerevisiae) (ENSG00000133488), score: 0.96 SEC16BSEC16 homolog B (S. cerevisiae) (ENSG00000120341), score: 0.72 SEC24ASEC24 family, member A (S. cerevisiae) (ENSG00000113615), score: 0.74 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (ENSG00000197249), score: 0.72 SERPINA10serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 (ENSG00000140093), score: 0.85 SERPINA11serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 (ENSG00000186910), score: 0.91 SERPINA4serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 (ENSG00000100665), score: 0.93 SERPINA6serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 (ENSG00000170099), score: 0.79 SERPINA7serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 (ENSG00000123561), score: 0.92 SERPINC1serpin peptidase inhibitor, clade C (antithrombin), member 1 (ENSG00000117601), score: 0.86 SERPIND1serpin peptidase inhibitor, clade D (heparin cofactor), member 1 (ENSG00000099937), score: 0.87 SHMT2serine hydroxymethyltransferase 2 (mitochondrial) (ENSG00000182199), score: 0.73 SIGMAR1sigma non-opioid intracellular receptor 1 (ENSG00000147955), score: 0.91 SKP2S-phase kinase-associated protein 2 (p45) (ENSG00000145604), score: 0.88 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (ENSG00000100652), score: 0.95 SLC16A11solute carrier family 16, member 11 (monocarboxylic acid transporter 11) (ENSG00000174326), score: 0.72 SLC16A2solute carrier family 16, member 2 (monocarboxylic acid transporter 8) (ENSG00000147100), score: 0.87 SLC17A2solute carrier family 17 (sodium phosphate), member 2 (ENSG00000112337), score: 0.9 SLC17A4solute carrier family 17 (sodium phosphate), member 4 (ENSG00000146039), score: 0.81 SLC17A9solute carrier family 17, member 9 (ENSG00000101194), score: 0.73 SLC19A3solute carrier family 19, member 3 (ENSG00000135917), score: 0.78 SLC22A1solute carrier family 22 (organic cation transporter), member 1 (ENSG00000175003), score: 0.91 SLC25A13solute carrier family 25, member 13 (citrin) (ENSG00000004864), score: 0.77 SLC25A15solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 (ENSG00000102743), score: 0.94 SLC25A36solute carrier family 25, member 36 (ENSG00000114120), score: -0.81 SLC25A38solute carrier family 25, member 38 (ENSG00000144659), score: 0.77 SLC25A47solute carrier family 25, member 47 (ENSG00000140107), score: 0.9 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (ENSG00000197496), score: 0.74 SLC2A2solute carrier family 2 (facilitated glucose transporter), member 2 (ENSG00000163581), score: 0.86 SLC30A10solute carrier family 30, member 10 (ENSG00000196660), score: 0.79 SLC35D1solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 (ENSG00000116704), score: 0.88 SLC38A4solute carrier family 38, member 4 (ENSG00000139209), score: 0.93 SLC39A11solute carrier family 39 (metal ion transporter), member 11 (ENSG00000133195), score: 0.77 SLC39A8solute carrier family 39 (zinc transporter), member 8 (ENSG00000138821), score: 0.73 SLC41A2solute carrier family 41, member 2 (ENSG00000136052), score: 0.83 SLC43A1solute carrier family 43, member 1 (ENSG00000149150), score: 0.86 SLC46A3solute carrier family 46, member 3 (ENSG00000139508), score: 0.78 SLC7A2solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 (ENSG00000003989), score: 0.77 SLCO1B1solute carrier organic anion transporter family, member 1B1 (ENSG00000134538), score: 0.92 SLCO1B3solute carrier organic anion transporter family, member 1B3 (ENSG00000111700), score: 1 SLPIsecretory leukocyte peptidase inhibitor (ENSG00000124107), score: 0.74 SMPDL3Asphingomyelin phosphodiesterase, acid-like 3A (ENSG00000172594), score: 0.75 SNTB1syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) (ENSG00000172164), score: 0.78 SOAT2sterol O-acyltransferase 2 (ENSG00000167780), score: 0.97 SORT1sortilin 1 (ENSG00000134243), score: -0.74 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.97 SPRYD4SPRY domain containing 4 (ENSG00000176422), score: 0.74 SRD5A2steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) (ENSG00000049319), score: 0.93 SRPRsignal recognition particle receptor (docking protein) (ENSG00000182934), score: 0.72 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (ENSG00000073849), score: 0.74 STAB2stabilin 2 (ENSG00000136011), score: 0.84 STK39serine threonine kinase 39 (ENSG00000198648), score: -0.77 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (ENSG00000173597), score: 0.82 SULT2A1sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 (ENSG00000105398), score: 0.89 TATtyrosine aminotransferase (ENSG00000198650), score: 0.9 TBC1D22BTBC1 domain family, member 22B (ENSG00000065491), score: -0.75 TDO2tryptophan 2,3-dioxygenase (ENSG00000151790), score: 0.82 TECTBtectorin beta (ENSG00000119913), score: 0.74 TFR2transferrin receptor 2 (ENSG00000106327), score: 0.83 THRSPthyroid hormone responsive (ENSG00000151365), score: 0.85 TM4SF4transmembrane 4 L six family member 4 (ENSG00000169903), score: 0.91 TM6SF2transmembrane 6 superfamily member 2 (ENSG00000213996), score: 0.77 TM7SF2transmembrane 7 superfamily member 2 (ENSG00000149809), score: 0.76 TMBIM6transmembrane BAX inhibitor motif containing 6 (ENSG00000139644), score: 0.73 TMEM195transmembrane protein 195 (ENSG00000187546), score: 0.92 TMEM97transmembrane protein 97 (ENSG00000109084), score: 0.74 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.78 TRIB3tribbles homolog 3 (Drosophila) (ENSG00000101255), score: 0.79 TRPM8transient receptor potential cation channel, subfamily M, member 8 (ENSG00000144481), score: 0.73 TSPAN3tetraspanin 3 (ENSG00000140391), score: -0.78 TTC39Ctetratricopeptide repeat domain 39C (ENSG00000168234), score: 0.8 TTPAtocopherol (alpha) transfer protein (ENSG00000137561), score: 0.92 TTPALtocopherol (alpha) transfer protein-like (ENSG00000124120), score: 0.82 TTRtransthyretin (ENSG00000118271), score: 0.81 UGP2UDP-glucose pyrophosphorylase 2 (ENSG00000169764), score: 0.76 UNC93Aunc-93 homolog A (C. elegans) (ENSG00000112494), score: 0.9 UROC1urocanase domain containing 1 (ENSG00000159650), score: 0.98 VWCEvon Willebrand factor C and EGF domains (ENSG00000167992), score: 0.97 XBP1X-box binding protein 1 (ENSG00000100219), score: 0.72 XDHxanthine dehydrogenase (ENSG00000158125), score: 0.88 ZNF512zinc finger protein 512 (ENSG00000243943), score: -0.76
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppa_lv_m_ca1 | ppa | lv | m | _ |
| ppa_lv_f_ca1 | ppa | lv | f | _ |
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| ptr_lv_m_ca1 | ptr | lv | m | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| mml_lv_m_ca1 | mml | lv | m | _ |