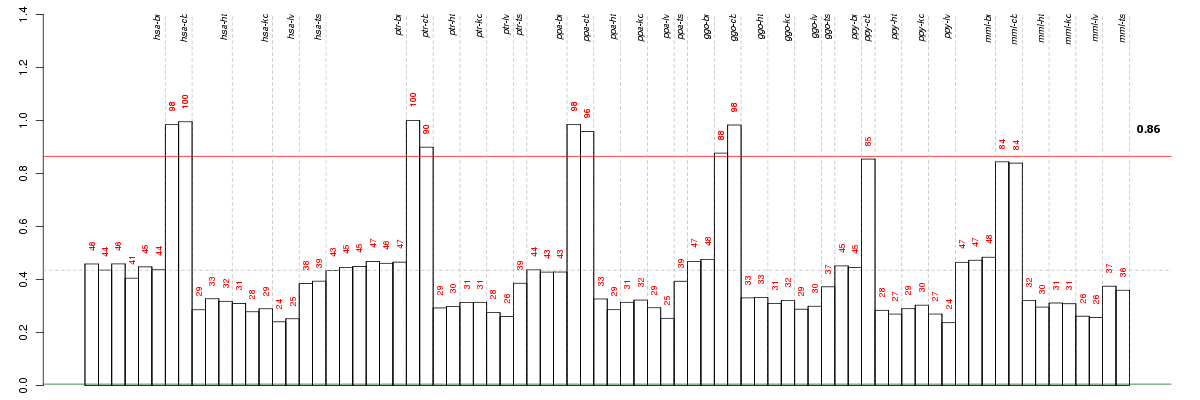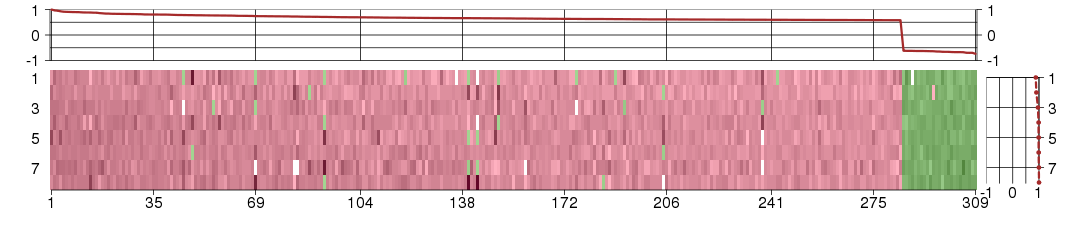

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The cellular synthesis of RNA on a template of DNA.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
cell surface receptor linked signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
enzyme linked receptor protein signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell, where the receptor possesses catalytic activity or is closely associated with an enzyme such as a protein kinase.
transmembrane receptor protein tyrosine kinase signaling pathway
The series of molecular signals generated as a consequence of a transmembrane receptor tyrosine kinase binding to its physiological ligand.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
brain development
The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The cellular synthesis of RNA on a template of DNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
brain development
The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.).
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
DNA binding
Any molecular function by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
glutamate receptor activity
Combining with glutamate to initiate a change in cell activity.
sequence-specific DNA binding
Interacting selectively and non-covalently with DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
| Id | Pvalue | ExpCount | Count | Size |
|---|---|---|---|---|
| miR-129/129-5p | 1.687e-03 | 7.965 | 25 | 275 |
ADAMTS16ADAM metallopeptidase with thrombospondin type 1 motif, 16 (ENSG00000145536), score: 0.62 ADARB1adenosine deaminase, RNA-specific, B1 (ENSG00000197381), score: 0.61 AMICA1adhesion molecule, interacts with CXADR antigen 1 (ENSG00000160593), score: 0.69 AMIGO3adhesion molecule with Ig-like domain 3 (ENSG00000176020), score: 0.63 ANKLE1ankyrin repeat and LEM domain containing 1 (ENSG00000160117), score: 0.85 ANKS6ankyrin repeat and sterile alpha motif domain containing 6 (ENSG00000165138), score: 0.68 ARMC8armadillo repeat containing 8 (ENSG00000114098), score: 0.6 ARPC2actin related protein 2/3 complex, subunit 2, 34kDa (ENSG00000163466), score: -0.63 ARSJarylsulfatase family, member J (ENSG00000180801), score: 0.59 ARVCFarmadillo repeat gene deleted in velocardiofacial syndrome (ENSG00000099889), score: 0.65 ATF7activating transcription factor 7 (ENSG00000170653), score: 0.68 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: -0.66 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (ENSG00000112182), score: 0.61 BAHCC1BAH domain and coiled-coil containing 1 (ENSG00000171282), score: 0.72 BANK1B-cell scaffold protein with ankyrin repeats 1 (ENSG00000153064), score: 0.72 BARHL1BarH-like homeobox 1 (ENSG00000125492), score: 0.86 BARHL2BarH-like homeobox 2 (ENSG00000143032), score: 0.91 BEST4bestrophin 4 (ENSG00000142959), score: 0.69 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.64 BPTFbromodomain PHD finger transcription factor (ENSG00000171634), score: 0.66 BTN2A1butyrophilin, subfamily 2, member A1 (ENSG00000112763), score: 0.71 C12orf51chromosome 12 open reading frame 51 (ENSG00000173064), score: 0.58 C14orf38chromosome 14 open reading frame 38 (ENSG00000151838), score: 0.58 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.75 C16orf11chromosome 16 open reading frame 11 (ENSG00000161992), score: 0.8 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: 0.61 C1orf127chromosome 1 open reading frame 127 (ENSG00000175262), score: 0.65 C1orf35chromosome 1 open reading frame 35 (ENSG00000143793), score: 0.6 C20orf117chromosome 20 open reading frame 117 (ENSG00000149639), score: 0.71 C6orf115chromosome 6 open reading frame 115 (ENSG00000146386), score: -0.67 C7orf16chromosome 7 open reading frame 16 (ENSG00000106341), score: 0.63 C8orf79chromosome 8 open reading frame 79 (ENSG00000170941), score: 0.66 CACNA1Acalcium channel, voltage-dependent, P/Q type, alpha 1A subunit (ENSG00000141837), score: 0.63 CAMK4calcium/calmodulin-dependent protein kinase IV (ENSG00000152495), score: 0.59 CAMKK2calcium/calmodulin-dependent protein kinase kinase 2, beta (ENSG00000110931), score: 0.6 CAPN10calpain 10 (ENSG00000142330), score: 0.58 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.83 CBLN3cerebellin 3 precursor (ENSG00000139899), score: 0.82 CCDC109Acoiled-coil domain containing 109A (ENSG00000156026), score: 0.7 CCDC129coiled-coil domain containing 129 (ENSG00000180347), score: 0.6 CCNJLcyclin J-like (ENSG00000135083), score: 0.73 CDC25Bcell division cycle 25 homolog B (S. pombe) (ENSG00000101224), score: 0.59 CDC42BPGCDC42 binding protein kinase gamma (DMPK-like) (ENSG00000171219), score: 0.65 CDH15cadherin 15, type 1, M-cadherin (myotubule) (ENSG00000129910), score: 0.9 CDH23cadherin-related 23 (ENSG00000107736), score: 0.61 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.73 CDKN1Bcyclin-dependent kinase inhibitor 1B (p27, Kip1) (ENSG00000111276), score: 0.59 CDONCdon homolog (mouse) (ENSG00000064309), score: 0.74 CDR2Lcerebellar degeneration-related protein 2-like (ENSG00000109089), score: 0.58 CECR2cat eye syndrome chromosome region, candidate 2 (ENSG00000099954), score: 0.71 CERKLceramide kinase-like (ENSG00000188452), score: 0.68 CHD7chromodomain helicase DNA binding protein 7 (ENSG00000171316), score: 0.8 CHN2chimerin (chimaerin) 2 (ENSG00000106069), score: 0.66 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.77 CLK4CDC-like kinase 4 (ENSG00000113240), score: 0.77 CLVS2clavesin 2 (ENSG00000146352), score: 0.6 CNOT2CCR4-NOT transcription complex, subunit 2 (ENSG00000111596), score: 0.7 CNPY1canopy 1 homolog (zebrafish) (ENSG00000146910), score: 0.82 CNTN6contactin 6 (ENSG00000134115), score: 0.61 COL13A1collagen, type XIII, alpha 1 (ENSG00000197467), score: 0.96 COL27A1collagen, type XXVII, alpha 1 (ENSG00000196739), score: 0.65 COL5A2collagen, type V, alpha 2 (ENSG00000204262), score: -0.67 COL8A2collagen, type VIII, alpha 2 (ENSG00000171812), score: 0.62 CPLX4complexin 4 (ENSG00000166569), score: 0.74 CRTAMcytotoxic and regulatory T cell molecule (ENSG00000109943), score: 0.95 CTCFCCCTC-binding factor (zinc finger protein) (ENSG00000102974), score: 0.62 CYB5Bcytochrome b5 type B (outer mitochondrial membrane) (ENSG00000103018), score: -0.63 DAB2IPDAB2 interacting protein (ENSG00000136848), score: 0.63 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (ENSG00000165617), score: 0.62 DCLRE1BDNA cross-link repair 1B (ENSG00000118655), score: 0.76 DCP1ADCP1 decapping enzyme homolog A (S. cerevisiae) (ENSG00000162290), score: 0.63 DGCR8DiGeorge syndrome critical region gene 8 (ENSG00000128191), score: 0.69 DGKDdiacylglycerol kinase, delta 130kDa (ENSG00000077044), score: 0.61 DGKGdiacylglycerol kinase, gamma 90kDa (ENSG00000058866), score: 0.59 DHRS13dehydrogenase/reductase (SDR family) member 13 (ENSG00000167536), score: 0.69 DNMT3ADNA (cytosine-5-)-methyltransferase 3 alpha (ENSG00000119772), score: 0.59 DPF3D4, zinc and double PHD fingers, family 3 (ENSG00000205683), score: 0.59 DUOX1dual oxidase 1 (ENSG00000137857), score: 0.78 ECE1endothelin converting enzyme 1 (ENSG00000117298), score: -0.67 EML1echinoderm microtubule associated protein like 1 (ENSG00000066629), score: -0.62 EN2engrailed homeobox 2 (ENSG00000164778), score: 0.87 EOMESeomesodermin (ENSG00000163508), score: 0.77 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: 0.62 EPC1enhancer of polycomb homolog 1 (Drosophila) (ENSG00000120616), score: 0.67 EPHB1EPH receptor B1 (ENSG00000154928), score: 0.63 ESPNLespin-like (ENSG00000144488), score: 0.63 ESYT3extended synaptotagmin-like protein 3 (ENSG00000158220), score: 0.71 ETV6ets variant 6 (ENSG00000139083), score: -0.64 EXPH5exophilin 5 (ENSG00000110723), score: 0.79 FAM110Bfamily with sequence similarity 110, member B (ENSG00000169122), score: 0.59 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.89 FBXO31F-box protein 31 (ENSG00000103264), score: 0.64 FGF17fibroblast growth factor 17 (ENSG00000158815), score: 0.74 FGF20fibroblast growth factor 20 (ENSG00000078579), score: 0.65 FGF3fibroblast growth factor 3 (ENSG00000186895), score: 0.71 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.83 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.68 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: 0.58 FLT3fms-related tyrosine kinase 3 (ENSG00000122025), score: 0.66 FOXN2forkhead box N2 (ENSG00000170802), score: 0.61 FSTL5follistatin-like 5 (ENSG00000168843), score: 0.72 FUT9fucosyltransferase 9 (alpha (1,3) fucosyltransferase) (ENSG00000172461), score: 0.63 FZD7frizzled homolog 7 (Drosophila) (ENSG00000155760), score: 0.75 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (ENSG00000119514), score: 0.59 GALNT5UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) (ENSG00000136542), score: 0.61 GALNT7UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (ENSG00000109586), score: 0.6 GDF10growth differentiation factor 10 (ENSG00000107623), score: 0.59 GIGYF1GRB10 interacting GYF protein 1 (ENSG00000146830), score: 0.66 GLCEglucuronic acid epimerase (ENSG00000138604), score: 0.68 GNG13guanine nucleotide binding protein (G protein), gamma 13 (ENSG00000127588), score: 0.62 GPATCH8G patch domain containing 8 (ENSG00000186566), score: 0.69 GPRIN3GPRIN family member 3 (ENSG00000185477), score: 0.77 GRIA4glutamate receptor, ionotrophic, AMPA 4 (ENSG00000152578), score: 0.58 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.73 GRID2IPglutamate receptor, ionotropic, delta 2 (Grid2) interacting protein (ENSG00000215045), score: 0.84 GRIN2Cglutamate receptor, ionotropic, N-methyl D-aspartate 2C (ENSG00000161509), score: 0.74 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.65 GRM4glutamate receptor, metabotropic 4 (ENSG00000124493), score: 0.82 HINFPhistone H4 transcription factor (ENSG00000172273), score: 0.7 HIPK1homeodomain interacting protein kinase 1 (ENSG00000163349), score: 0.59 IFFO2intermediate filament family orphan 2 (ENSG00000169991), score: 0.73 IGFBP2insulin-like growth factor binding protein 2, 36kDa (ENSG00000115457), score: -0.7 IL16interleukin 16 (lymphocyte chemoattractant factor) (ENSG00000172349), score: 0.8 INCENPinner centromere protein antigens 135/155kDa (ENSG00000149503), score: 0.68 INO80INO80 homolog (S. cerevisiae) (ENSG00000128908), score: 0.67 IP6K2inositol hexakisphosphate kinase 2 (ENSG00000068745), score: 0.7 JMJD1Cjumonji domain containing 1C (ENSG00000171988), score: 0.67 KCNAB3potassium voltage-gated channel, shaker-related subfamily, beta member 3 (ENSG00000170049), score: 0.61 KCNC1potassium voltage-gated channel, Shaw-related subfamily, member 1 (ENSG00000129159), score: 0.6 KCND1potassium voltage-gated channel, Shal-related subfamily, member 1 (ENSG00000102057), score: 0.63 KCNK10potassium channel, subfamily K, member 10 (ENSG00000100433), score: 0.62 KCNK9potassium channel, subfamily K, member 9 (ENSG00000169427), score: 0.74 KCNRGpotassium channel regulator (ENSG00000198553), score: 0.68 KCTD2potassium channel tetramerisation domain containing 2 (ENSG00000180901), score: 0.64 KDM4Clysine (K)-specific demethylase 4C (ENSG00000107077), score: 0.82 KIAA0182KIAA0182 (ENSG00000131149), score: 0.67 KIAA0247KIAA0247 (ENSG00000100647), score: 0.63 KIAA0802KIAA0802 (ENSG00000168502), score: 0.78 KIAA0907KIAA0907 (ENSG00000132680), score: 0.59 KIAA1614KIAA1614 (ENSG00000135835), score: 0.6 KLHDC8Akelch domain containing 8A (ENSG00000162873), score: 0.59 KLHL3kelch-like 3 (Drosophila) (ENSG00000146021), score: 0.61 KRT39keratin 39 (ENSG00000196859), score: 0.66 KRT40keratin 40 (ENSG00000204889), score: 0.66 LASS5LAG1 homolog, ceramide synthase 5 (ENSG00000139624), score: 0.58 LBX1ladybird homeobox 1 (ENSG00000138136), score: 0.79 LDB2LIM domain binding 2 (ENSG00000169744), score: -0.74 LDLRAD3low density lipoprotein receptor class A domain containing 3 (ENSG00000179241), score: 0.62 LDLRAP1low density lipoprotein receptor adaptor protein 1 (ENSG00000157978), score: 0.64 LENEPlens epithelial protein (ENSG00000163352), score: 0.62 LHX5LIM homeobox 5 (ENSG00000089116), score: 0.75 LMNB1lamin B1 (ENSG00000113368), score: 0.59 LMO7LIM domain 7 (ENSG00000136153), score: -0.66 LOC100131509similar to inward rectifying K+ channel negative regulator Kir2.2v (ENSG00000184185), score: 0.6 LOC100294337hypothetical protein LOC100294337 (ENSG00000120071), score: 0.6 LOC146429putative solute carrier family 22 member ENSG00000182157 (ENSG00000182157), score: 0.93 LONRF3LON peptidase N-terminal domain and ring finger 3 (ENSG00000175556), score: 0.61 LRCH1leucine-rich repeats and calponin homology (CH) domain containing 1 (ENSG00000136141), score: 0.71 LRRC16Bleucine rich repeat containing 16B (ENSG00000186648), score: 0.62 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.73 LRRC38leucine rich repeat containing 38 (ENSG00000162494), score: 0.65 LRRC3Bleucine rich repeat containing 3B (ENSG00000179796), score: 0.61 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.9 MAML1mastermind-like 1 (Drosophila) (ENSG00000161021), score: 0.75 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.77 MAMSTRMEF2 activating motif and SAP domain containing transcriptional regulator (ENSG00000176909), score: 0.63 MAXMYC associated factor X (ENSG00000125952), score: 0.72 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.84 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.7 MLLmyeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) (ENSG00000118058), score: 0.66 MMP24matrix metallopeptidase 24 (membrane-inserted) (ENSG00000125966), score: 0.62 MOV10Mov10, Moloney leukemia virus 10, homolog (mouse) (ENSG00000155363), score: -0.63 MPP3membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) (ENSG00000161647), score: 0.64 MRGPRDMAS-related GPR, member D (ENSG00000172938), score: 0.65 MRPL24mitochondrial ribosomal protein L24 (ENSG00000143314), score: -0.65 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: -0.68 MSX2msh homeobox 2 (ENSG00000120149), score: 0.63 MYO1Emyosin IE (ENSG00000157483), score: -0.63 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.7 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (ENSG00000166886), score: 0.64 NHLH2nescient helix loop helix 2 (ENSG00000177551), score: 0.91 NKAIN1Na+/K+ transporting ATPase interacting 1 (ENSG00000084628), score: 0.73 NKX6-3NK6 homeobox 3 (ENSG00000165066), score: 0.88 NODALnodal homolog (mouse) (ENSG00000156574), score: 0.72 NPFFneuropeptide FF-amide peptide precursor (ENSG00000139574), score: 0.61 NPR2natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) (ENSG00000159899), score: 0.59 NRG2neuregulin 2 (ENSG00000158458), score: 0.66 NRIP2nuclear receptor interacting protein 2 (ENSG00000053702), score: 0.78 NYNRINNYN domain and retroviral integrase containing (ENSG00000205978), score: 0.71 ODZ1odz, odd Oz/ten-m homolog 1(Drosophila) (ENSG00000009694), score: 0.8 OGFRL1opioid growth factor receptor-like 1 (ENSG00000119900), score: 0.6 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: 0.67 OTX2orthodenticle homeobox 2 (ENSG00000165588), score: 0.77 PAG1phosphoprotein associated with glycosphingolipid microdomains 1 (ENSG00000076641), score: 0.63 PAX6paired box 6 (ENSG00000007372), score: 0.75 PAXIP1PAX interacting (with transcription-activation domain) protein 1 (ENSG00000157212), score: 0.75 PCBP3poly(rC) binding protein 3 (ENSG00000183570), score: 0.63 PCBP4poly(rC) binding protein 4 (ENSG00000090097), score: 0.58 PCIF1PDX1 C-terminal inhibiting factor 1 (ENSG00000100982), score: 0.6 PELI2pellino homolog 2 (Drosophila) (ENSG00000139946), score: 0.61 PHF2PHD finger protein 2 (ENSG00000197724), score: 0.67 PHIPpleckstrin homology domain interacting protein (ENSG00000146247), score: 0.59 PIAS3protein inhibitor of activated STAT, 3 (ENSG00000131788), score: 0.59 PIGPphosphatidylinositol glycan anchor biosynthesis, class P (ENSG00000185808), score: -0.62 PISDphosphatidylserine decarboxylase (ENSG00000241878), score: 0.8 PKIBprotein kinase (cAMP-dependent, catalytic) inhibitor beta (ENSG00000135549), score: 0.88 PLCB4phospholipase C, beta 4 (ENSG00000101333), score: 0.61 PLD5phospholipase D family, member 5 (ENSG00000180287), score: 0.6 PLK2polo-like kinase 2 (ENSG00000145632), score: -0.65 PLXNA3plexin A3 (ENSG00000130827), score: 0.6 PLXND1plexin D1 (ENSG00000004399), score: -0.67 POLBpolymerase (DNA directed), beta (ENSG00000070501), score: 0.6 POLE4polymerase (DNA-directed), epsilon 4 (p12 subunit) (ENSG00000115350), score: -0.64 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.73 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (ENSG00000101161), score: 0.63 PTCH1patched 1 (ENSG00000185920), score: 0.66 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.77 PWWP2APWWP domain containing 2A (ENSG00000170234), score: 0.7 PYDC1PYD (pyrin domain) containing 1 (ENSG00000169900), score: 0.83 PYGO1pygopus homolog 1 (Drosophila) (ENSG00000171016), score: 0.6 QSOX2quiescin Q6 sulfhydryl oxidase 2 (ENSG00000165661), score: 0.67 RAB37RAB37, member RAS oncogene family (ENSG00000172794), score: 0.69 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.87 REV3LREV3-like, catalytic subunit of DNA polymerase zeta (yeast) (ENSG00000009413), score: 0.67 RFTN1raftlin, lipid raft linker 1 (ENSG00000131378), score: -0.7 RIT2Ras-like without CAAX 2 (ENSG00000152214), score: 0.61 RMND5Arequired for meiotic nuclear division 5 homolog A (S. cerevisiae) (ENSG00000153561), score: 0.67 RNF146ring finger protein 146 (ENSG00000118518), score: 0.62 RPS6KA5ribosomal protein S6 kinase, 90kDa, polypeptide 5 (ENSG00000100784), score: 0.58 RTEL1regulator of telomere elongation helicase 1 (ENSG00000026036), score: 0.63 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: 0.68 SBK1SH3-binding domain kinase 1 (ENSG00000188322), score: 0.6 SCGNsecretagogin, EF-hand calcium binding protein (ENSG00000079689), score: 0.76 SEL1L3sel-1 suppressor of lin-12-like 3 (C. elegans) (ENSG00000091490), score: 0.61 SETD5SET domain containing 5 (ENSG00000168137), score: 0.66 SGK223homolog of rat pragma of Rnd2 (ENSG00000182319), score: 0.69 SHBSrc homology 2 domain containing adaptor protein B (ENSG00000107338), score: -0.7 SKIv-ski sarcoma viral oncogene homolog (avian) (ENSG00000157933), score: 0.67 SKOR1SKI family transcriptional corepressor 1 (ENSG00000188779), score: 0.64 SLC1A6solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 (ENSG00000105143), score: 0.61 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.59 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.9 SLC36A1solute carrier family 36 (proton/amino acid symporter), member 1 (ENSG00000123643), score: 0.59 SLC9A5solute carrier family 9 (sodium/hydrogen exchanger), member 5 (ENSG00000135740), score: 0.59 SNED1sushi, nidogen and EGF-like domains 1 (ENSG00000162804), score: 0.6 SNRKSNF related kinase (ENSG00000163788), score: 0.72 SNX25sorting nexin 25 (ENSG00000109762), score: 0.65 SOCS5suppressor of cytokine signaling 5 (ENSG00000171150), score: 0.6 SP4Sp4 transcription factor (ENSG00000105866), score: 0.68 SPHKAPSPHK1 interactor, AKAP domain containing (ENSG00000153820), score: 0.61 SPINK6serine peptidase inhibitor, Kazal type 6 (ENSG00000178172), score: 0.78 SPTBN5spectrin, beta, non-erythrocytic 5 (ENSG00000137877), score: 0.76 SRRM4serine/arginine repetitive matrix 4 (ENSG00000139767), score: 0.58 ST8SIA1ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 (ENSG00000111728), score: 0.58 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.72 STAT5Bsignal transducer and activator of transcription 5B (ENSG00000173757), score: 0.59 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (ENSG00000166888), score: -0.66 SYT2synaptotagmin II (ENSG00000143858), score: 0.66 TAS2R3taste receptor, type 2, member 3 (ENSG00000127362), score: 0.76 TFAP2Etranscription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) (ENSG00000116819), score: 0.76 TGM1transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) (ENSG00000092295), score: 0.8 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.77 TIMP4TIMP metallopeptidase inhibitor 4 (ENSG00000157150), score: 0.6 TLL1tolloid-like 1 (ENSG00000038295), score: 0.81 TMC2transmembrane channel-like 2 (ENSG00000149488), score: 0.88 TMEM145transmembrane protein 145 (ENSG00000167619), score: 0.58 TMEM163transmembrane protein 163 (ENSG00000152128), score: 0.59 TNFRSF25tumor necrosis factor receptor superfamily, member 25 (ENSG00000215788), score: 0.61 TNK1tyrosine kinase, non-receptor, 1 (ENSG00000174292), score: 0.65 TP73tumor protein p73 (ENSG00000078900), score: 0.83 TPRNtaperin (ENSG00000176058), score: 0.6 TRIM11tripartite motif-containing 11 (ENSG00000154370), score: 0.82 TRIM17tripartite motif-containing 17 (ENSG00000162931), score: 0.66 TRIM32tripartite motif-containing 32 (ENSG00000119401), score: 0.6 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.83 TRIM7tripartite motif-containing 7 (ENSG00000146054), score: 0.69 TRIM72tripartite motif-containing 72 (ENSG00000177238), score: 0.63 UBASH3Bubiquitin associated and SH3 domain containing B (ENSG00000154127), score: 0.66 UPF0639UPF0639 protein (ENSG00000175985), score: 0.61 USTuronyl-2-sulfotransferase (ENSG00000111962), score: 0.66 VAT1Lvesicle amine transport protein 1 homolog (T. californica)-like (ENSG00000171724), score: 0.61 VAX2ventral anterior homeobox 2 (ENSG00000116035), score: 0.63 VSX1visual system homeobox 1 (ENSG00000100987), score: 1 VWA5Avon Willebrand factor A domain containing 5A (ENSG00000110002), score: -0.62 WSCD2WSC domain containing 2 (ENSG00000075035), score: 0.64 WWC2WW and C2 domain containing 2 (ENSG00000151718), score: -0.63 XKR7XK, Kell blood group complex subunit-related family, member 7 (ENSG00000101321), score: 0.8 YTHDF1YTH domain family, member 1 (ENSG00000149658), score: 0.66 ZBTB34zinc finger and BTB domain containing 34 (ENSG00000177125), score: 0.59 ZFHX3zinc finger homeobox 3 (ENSG00000140836), score: -0.62 ZFP112zinc finger protein 112 homolog (mouse) (ENSG00000062370), score: 0.59 ZFPM2zinc finger protein, multitype 2 (ENSG00000169946), score: 0.66 ZFYVE28zinc finger, FYVE domain containing 28 (ENSG00000159733), score: 0.6 ZIC2Zic family member 2 (odd-paired homolog, Drosophila) (ENSG00000043355), score: 0.79 ZIC4Zic family member 4 (ENSG00000174963), score: 0.9 ZIC5Zic family member 5 (odd-paired homolog, Drosophila) (ENSG00000139800), score: 0.81 ZNF157zinc finger protein 157 (ENSG00000147117), score: 0.7 ZNF238zinc finger protein 238 (ENSG00000179456), score: 0.65 ZNF273zinc finger protein 273 (ENSG00000198039), score: 0.58 ZNF296zinc finger protein 296 (ENSG00000170684), score: 0.76 ZNF304zinc finger protein 304 (ENSG00000131845), score: 0.61 ZNF333zinc finger protein 333 (ENSG00000160961), score: 0.59 ZNF34zinc finger protein 34 (ENSG00000196378), score: 0.67 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.8 ZNF589zinc finger protein 589 (ENSG00000164048), score: 0.6 ZNF606zinc finger protein 606 (ENSG00000166704), score: 0.63 ZNF671zinc finger protein 671 (ENSG00000083814), score: 0.69 ZNF772zinc finger protein 772 (ENSG00000197128), score: 0.61
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| ptr_cb_f_ca1 | ptr | cb | f | _ |
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| hsa_cb_m_ca1 | hsa | cb | m | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| hsa_cb_f_ca1 | hsa | cb | f | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |