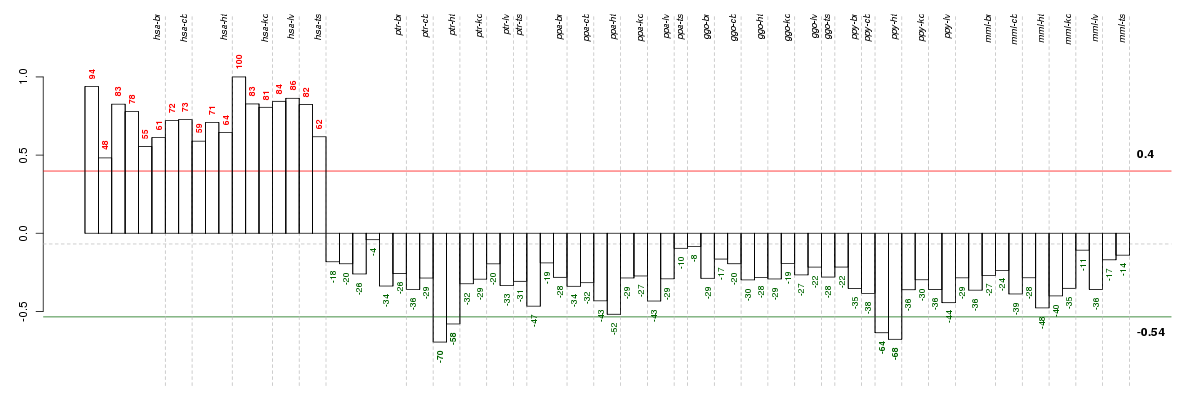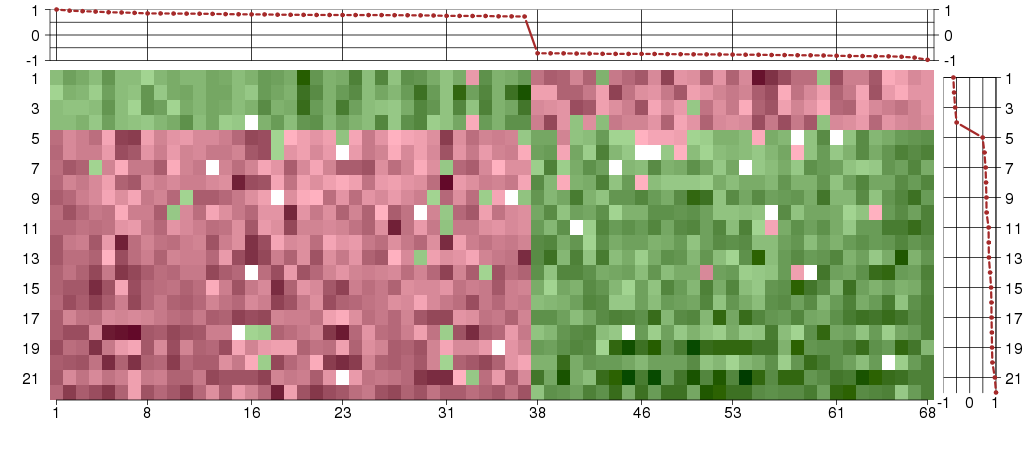

Under-expression is coded with green,
over-expression with red color.



AIDAaxin interactor, dorsalization associated (ENSG00000186063), score: -0.83 AKTIPAKT interacting protein (ENSG00000166971), score: -0.75 ATG10ATG10 autophagy related 10 homolog (S. cerevisiae) (ENSG00000152348), score: 0.75 BAT5HLA-B associated transcript 5 (ENSG00000204422), score: 1 C12orf52chromosome 12 open reading frame 52 (ENSG00000139405), score: 0.75 C2orf49chromosome 2 open reading frame 49 (ENSG00000135974), score: -0.82 C3orf16chromosome 3 open reading frame 16 (ENSG00000206199), score: 0.81 C5orf45chromosome 5 open reading frame 45 (ENSG00000161010), score: 0.85 C8orf38chromosome 8 open reading frame 38 (ENSG00000156170), score: -0.79 CD72CD72 molecule (ENSG00000137101), score: 0.82 CDC26cell division cycle 26 homolog (S. cerevisiae) (ENSG00000176386), score: -0.83 CITED4Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 (ENSG00000179862), score: 0.78 CLCC1chloride channel CLIC-like 1 (ENSG00000121940), score: -0.76 CLUL1clusterin-like 1 (retinal) (ENSG00000079101), score: 0.75 CNIH4cornichon homolog 4 (Drosophila) (ENSG00000143771), score: -0.72 COMMD2COMM domain containing 2 (ENSG00000114744), score: -0.76 CRIPTcysteine-rich PDZ-binding protein (ENSG00000119878), score: -0.79 DLEC1deleted in lung and esophageal cancer 1 (ENSG00000008226), score: 0.79 DPH1DPH1 homolog (S. cerevisiae) (ENSG00000108963), score: 0.78 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (ENSG00000105865), score: -0.72 EEF1Geukaryotic translation elongation factor 1 gamma (ENSG00000149016), score: 0.95 ELAC2elaC homolog 2 (E. coli) (ENSG00000006744), score: 0.78 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.73 EXOGendo/exonuclease (5'-3'), endonuclease G-like (ENSG00000157036), score: -0.78 FAM103A1family with sequence similarity 103, member A1 (ENSG00000169612), score: -0.84 FAM119Afamily with sequence similarity 119, member A (ENSG00000144401), score: -0.77 FAM156Afamily with sequence similarity 156, member A (ENSG00000182646), score: -0.8 GPR141G protein-coupled receptor 141 (ENSG00000187037), score: 0.81 GUK1guanylate kinase 1 (ENSG00000143774), score: 0.76 IL9Rinterleukin 9 receptor (ENSG00000124334), score: 0.92 LOC100133770hypothetical protein LOC100133770 (ENSG00000069329), score: -0.72 MFN1mitofusin 1 (ENSG00000171109), score: -0.77 MIFmacrophage migration inhibitory factor (glycosylation-inhibiting factor) (ENSG00000133460), score: 0.84 MLST8MTOR associated protein, LST8 homolog (S. cerevisiae) (ENSG00000167965), score: 0.77 MNAT1menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) (ENSG00000020426), score: -0.74 MYL5myosin, light chain 5, regulatory (ENSG00000215375), score: 0.85 NDOR1NADPH dependent diflavin oxidoreductase 1 (ENSG00000188566), score: 0.79 NOL8nucleolar protein 8 (ENSG00000198000), score: -0.77 NPBneuropeptide B (ENSG00000183979), score: 0.84 NPRL3nitrogen permease regulator-like 3 (S. cerevisiae) (ENSG00000103148), score: 0.77 NT5C35'-nucleotidase, cytosolic III (ENSG00000122643), score: -0.74 OTUD6BOTU domain containing 6B (ENSG00000155100), score: -0.74 PIGCphosphatidylinositol glycan anchor biosynthesis, class C (ENSG00000135845), score: -0.74 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (ENSG00000171608), score: 0.81 PRICKLE4prickle homolog 4 (Drosophila) (ENSG00000124593), score: 0.75 PTCD2pentatricopeptide repeat domain 2 (ENSG00000049883), score: -0.73 RBM8ARNA binding motif protein 8A (ENSG00000131795), score: -0.89 RSPO1R-spondin homolog (Xenopus laevis) (ENSG00000169218), score: 0.79 SCNN1Dsodium channel, nonvoltage-gated 1, delta (ENSG00000162572), score: 0.83 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.76 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.87 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.89 SNX13sorting nexin 13 (ENSG00000071189), score: -0.81 SNX22sorting nexin 22 (ENSG00000157734), score: 0.93 TH1LTH1-like (Drosophila) (ENSG00000101158), score: 0.78 THUMPD1THUMP domain containing 1 (ENSG00000066654), score: -0.74 TMEM199transmembrane protein 199 (ENSG00000244045), score: -0.85 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.8 TRIM69tripartite motif-containing 69 (ENSG00000185880), score: 0.78 TRNT1tRNA nucleotidyl transferase, CCA-adding, 1 (ENSG00000072756), score: -0.75 TSPAN16tetraspanin 16 (ENSG00000130167), score: 0.83 UTS2urotensin 2 (ENSG00000049247), score: 0.88 WDR83WD repeat domain 83 (ENSG00000123154), score: 0.72 ZACNzinc activated ligand-gated ion channel (ENSG00000186919), score: 0.73 ZC3H6zinc finger CCCH-type containing 6 (ENSG00000188177), score: -0.8 ZNF581zinc finger protein 581 (ENSG00000171425), score: 0.79
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_ht_m_ca1 | ptr | ht | m | _ |
| ppy_ht_f_ca1 | ppy | ht | f | _ |
| ppy_ht_m_ca1 | ppy | ht | m | _ |
| ptr_ht_f_ca1 | ptr | ht | f | _ |
| hsa_br_m2_ca1 | hsa | br | m | 2 |
| hsa_br_m1_ca1 | hsa | br | m | 1 |
| hsa_ht_m2_ca1 | hsa | ht | m | 2 |
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_ts_m2_ca1 | hsa | ts | m | 2 |
| hsa_ht_f_ca1 | hsa | ht | f | _ |
| hsa_ht_m1_ca1 | hsa | ht | m | 1 |
| hsa_cb_m_ca1 | hsa | cb | m | _ |
| hsa_cb_f_ca1 | hsa | cb | f | _ |
| hsa_br_m6_ca1 | hsa | br | m | 6 |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| hsa_ts_m1_ca1 | hsa | ts | m | 1 |
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |