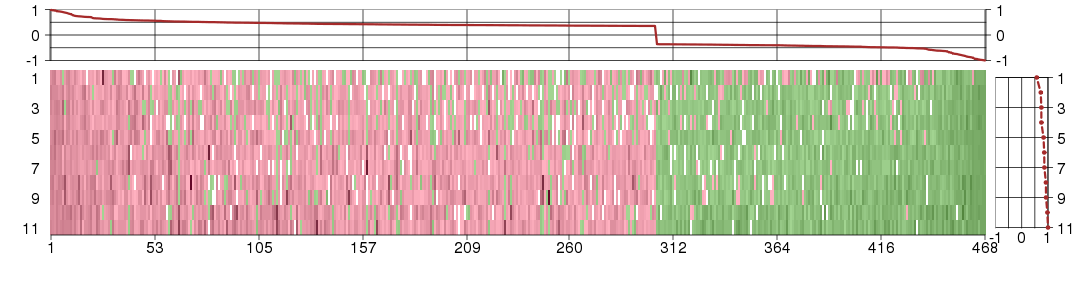

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
DNA metabolic process
Any cellular metabolic process involving deoxyribonucleic acid. This is one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides.
DNA repair
The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
response to DNA damage stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
DNA metabolic process
Any cellular metabolic process involving deoxyribonucleic acid. This is one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides.
DNA repair
The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway.


AAGABalpha- and gamma-adaptin binding protein (ENSG00000103591), score: -0.46 ABCC1ATP-binding cassette, sub-family C (CFTR/MRP), member 1 (ENSG00000103222), score: 0.54 ACADVLacyl-CoA dehydrogenase, very long chain (ENSG00000072778), score: 0.36 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: -0.51 ADH7alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide (ENSG00000196344), score: 0.88 ADPGKADP-dependent glucokinase (ENSG00000159322), score: 0.36 AGPSalkylglycerone phosphate synthase (ENSG00000018510), score: -0.37 AHCTF1AT hook containing transcription factor 1 (ENSG00000153207), score: -0.53 AHSA2AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) (ENSG00000173209), score: -0.63 AKIRIN2akirin 2 (ENSG00000135334), score: -0.36 ALDH3A1aldehyde dehydrogenase 3 family, member A1 (ENSG00000108602), score: 0.42 ALOX12arachidonate 12-lipoxygenase (ENSG00000108839), score: -0.4 AMICA1adhesion molecule, interacts with CXADR antigen 1 (ENSG00000160593), score: 0.38 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.53 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: -0.4 ANKRD54ankyrin repeat domain 54 (ENSG00000100124), score: -0.77 ANO10anoctamin 10 (ENSG00000160746), score: -0.37 ANO7anoctamin 7 (ENSG00000146205), score: 0.39 ANP32Bacidic (leucine-rich) nuclear phosphoprotein 32 family, member B (ENSG00000136938), score: -0.96 APTXaprataxin (ENSG00000137074), score: 0.66 ARCN1archain 1 (ENSG00000095139), score: -0.48 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (ENSG00000101199), score: 0.45 ARHGEF10Rho guanine nucleotide exchange factor (GEF) 10 (ENSG00000104728), score: 0.36 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: 0.39 ATPAF2ATP synthase mitochondrial F1 complex assembly factor 2 (ENSG00000171953), score: 0.36 BAXBCL2-associated X protein (ENSG00000087088), score: 0.41 BBS4Bardet-Biedl syndrome 4 (ENSG00000140463), score: 0.43 BCS1LBCS1-like (S. cerevisiae) (ENSG00000074582), score: 0.39 BEND5BEN domain containing 5 (ENSG00000162373), score: 0.5 BEST1bestrophin 1 (ENSG00000167995), score: 0.53 BEST3bestrophin 3 (ENSG00000127325), score: 0.37 BFARbifunctional apoptosis regulator (ENSG00000103429), score: 0.51 BNIPLBCL2/adenovirus E1B 19kD interacting protein like (ENSG00000163141), score: 0.38 BTN3A1butyrophilin, subfamily 3, member A1 (ENSG00000026950), score: 0.53 BUB1Bbudding uninhibited by benzimidazoles 1 homolog beta (yeast) (ENSG00000156970), score: 0.57 C10orf128chromosome 10 open reading frame 128 (ENSG00000204161), score: 0.39 C10orf46chromosome 10 open reading frame 46 (ENSG00000151893), score: -0.42 C14orf143chromosome 14 open reading frame 143 (ENSG00000140025), score: -0.48 C14orf45chromosome 14 open reading frame 45 (ENSG00000119636), score: -0.38 C17orf90chromosome 17 open reading frame 90 (ENSG00000204237), score: 0.48 C1orf69chromosome 1 open reading frame 69 (ENSG00000181873), score: 0.37 C1orf89chromosome 1 open reading frame 89 (ENSG00000132881), score: -0.5 C1QTNF5C1q and tumor necrosis factor related protein 5 (ENSG00000235718), score: 0.55 C1RLcomplement component 1, r subcomponent-like (ENSG00000139178), score: 0.43 C20orf152chromosome 20 open reading frame 152 (ENSG00000149646), score: 0.51 C21orf45chromosome 21 open reading frame 45 (ENSG00000159055), score: -0.41 C21orf56chromosome 21 open reading frame 56 (ENSG00000160284), score: -0.36 C21orf7chromosome 21 open reading frame 7 (ENSG00000156265), score: 0.55 C6orf201chromosome 6 open reading frame 201 (ENSG00000185689), score: -0.37 C6orf72chromosome 6 open reading frame 72 (ENSG00000055211), score: 0.39 C6orf89chromosome 6 open reading frame 89 (ENSG00000198663), score: -0.4 C8orf45chromosome 8 open reading frame 45 (ENSG00000178460), score: 0.93 C9orf173chromosome 9 open reading frame 173 (ENSG00000197768), score: 0.36 CAB39Lcalcium binding protein 39-like (ENSG00000102547), score: -0.49 CABP7calcium binding protein 7 (ENSG00000100314), score: 0.42 CACNA2D4calcium channel, voltage-dependent, alpha 2/delta subunit 4 (ENSG00000151062), score: 0.92 CALML6calmodulin-like 6 (ENSG00000169885), score: 0.48 CAPN12calpain 12 (ENSG00000182472), score: -0.41 CAPSLcalcyphosine-like (ENSG00000152611), score: 0.43 CARD6caspase recruitment domain family, member 6 (ENSG00000132357), score: 0.56 CASP5caspase 5, apoptosis-related cysteine peptidase (ENSG00000137757), score: 0.4 CBFBcore-binding factor, beta subunit (ENSG00000067955), score: 0.51 CCDC106coiled-coil domain containing 106 (ENSG00000173581), score: -0.48 CCDC107coiled-coil domain containing 107 (ENSG00000159884), score: -0.98 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: 0.45 CCDC12coiled-coil domain containing 12 (ENSG00000160799), score: 0.44 CCDC137coiled-coil domain containing 137 (ENSG00000185298), score: 0.6 CCDC51coiled-coil domain containing 51 (ENSG00000164051), score: 0.52 CCDC6coiled-coil domain containing 6 (ENSG00000108091), score: -0.37 CCDC90Bcoiled-coil domain containing 90B (ENSG00000137500), score: -0.4 CCL11chemokine (C-C motif) ligand 11 (ENSG00000172156), score: 0.44 CCT6Achaperonin containing TCP1, subunit 6A (zeta 1) (ENSG00000146731), score: -0.38 CD3EAPCD3e molecule, epsilon associated protein (ENSG00000117877), score: 0.49 CD3GCD3g molecule, gamma (CD3-TCR complex) (ENSG00000160654), score: 0.36 CDC26cell division cycle 26 homolog (S. cerevisiae) (ENSG00000176386), score: 0.44 CDK4cyclin-dependent kinase 4 (ENSG00000135446), score: 0.38 CDK5RAP3CDK5 regulatory subunit associated protein 3 (ENSG00000108465), score: 0.45 CDSNcorneodesmosin (ENSG00000204539), score: 0.37 CEACAM19carcinoembryonic antigen-related cell adhesion molecule 19 (ENSG00000186567), score: 0.37 CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: -0.49 CENPAcentromere protein A (ENSG00000115163), score: -0.42 CGGBP1CGG triplet repeat binding protein 1 (ENSG00000163320), score: -0.42 CHEK1CHK1 checkpoint homolog (S. pombe) (ENSG00000149554), score: 0.4 CHIT1chitinase 1 (chitotriosidase) (ENSG00000133063), score: 0.56 CLDN6claudin 6 (ENSG00000184697), score: 0.9 CLEC4FC-type lectin domain family 4, member F (ENSG00000152672), score: 0.44 CNPY4canopy 4 homolog (zebrafish) (ENSG00000166997), score: 0.57 COG4component of oligomeric golgi complex 4 (ENSG00000103051), score: 0.39 COL8A1collagen, type VIII, alpha 1 (ENSG00000144810), score: 0.57 COLQcollagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase (ENSG00000206561), score: 0.37 COMMD10COMM domain containing 10 (ENSG00000145781), score: 0.41 COX18COX18 cytochrome c oxidase assembly homolog (S. cerevisiae) (ENSG00000163626), score: -0.4 CPSF4Lcleavage and polyadenylation specific factor 4-like (ENSG00000187959), score: 0.86 CUL1cullin 1 (ENSG00000055130), score: -0.97 CUX1cut-like homeobox 1 (ENSG00000160967), score: -0.45 CWF19L1CWF19-like 1, cell cycle control (S. pombe) (ENSG00000095485), score: -0.49 CXCL13chemokine (C-X-C motif) ligand 13 (ENSG00000156234), score: 0.58 CYP2W1cytochrome P450, family 2, subfamily W, polypeptide 1 (ENSG00000073067), score: 0.52 DCAKDdephospho-CoA kinase domain containing (ENSG00000172992), score: 0.41 DCTdopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) (ENSG00000080166), score: 0.72 DDB2damage-specific DNA binding protein 2, 48kDa (ENSG00000134574), score: 0.65 DDI2DNA-damage inducible 1 homolog 2 (S. cerevisiae) (ENSG00000197312), score: -0.38 DDX55DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 (ENSG00000111364), score: 0.54 DEM1defects in morphology 1 homolog (S. cerevisiae) (ENSG00000164002), score: 0.38 DET1de-etiolated homolog 1 (Arabidopsis) (ENSG00000140543), score: 0.41 DNAJC17DnaJ (Hsp40) homolog, subfamily C, member 17 (ENSG00000104129), score: 0.43 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: -0.37 DNTTdeoxynucleotidyltransferase, terminal (ENSG00000107447), score: 0.5 DOCK11dedicator of cytokinesis 11 (ENSG00000147251), score: 0.39 DOK3docking protein 3 (ENSG00000146094), score: 0.43 DPH5DPH5 homolog (S. cerevisiae) (ENSG00000117543), score: 0.47 DVL3dishevelled, dsh homolog 3 (Drosophila) (ENSG00000161202), score: 0.58 DYNLRB2dynein, light chain, roadblock-type 2 (ENSG00000168589), score: 0.38 DYRK4dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 (ENSG00000010219), score: -0.38 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.4 E2F6E2F transcription factor 6 (ENSG00000169016), score: 0.5 EARS2glutamyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000103356), score: -0.37 EIF3Leukaryotic translation initiation factor 3, subunit L (ENSG00000100129), score: 0.36 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.56 ENAMenamelin (ENSG00000132464), score: 0.4 EPC1enhancer of polycomb homolog 1 (Drosophila) (ENSG00000120616), score: 0.39 EPT1ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) (ENSG00000138018), score: -0.63 ERCC1excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) (ENSG00000012061), score: 0.73 EVC2Ellis van Creveld syndrome 2 (ENSG00000173040), score: 0.37 EXOC2exocyst complex component 2 (ENSG00000112685), score: -0.4 FABP9fatty acid binding protein 9, testis (ENSG00000205186), score: 0.46 FAM120AOSfamily with sequence similarity 120A opposite strand (ENSG00000188938), score: -0.45 FAM122Cfamily with sequence similarity 122C (ENSG00000156500), score: 0.6 FAM134Cfamily with sequence similarity 134, member C (ENSG00000141699), score: -0.45 FAM168Afamily with sequence similarity 168, member A (ENSG00000054965), score: -0.38 FAM18Afamily with sequence similarity 18, member A (ENSG00000166676), score: -0.49 FAM193Bfamily with sequence similarity 193, member B (ENSG00000146067), score: 0.47 FAM59Bfamily with sequence similarity 59, member B (ENSG00000157833), score: 0.65 FAM64Afamily with sequence similarity 64, member A (ENSG00000129195), score: 0.4 FAM89Afamily with sequence similarity 89, member A (ENSG00000182118), score: 0.39 FAM89Bfamily with sequence similarity 89, member B (ENSG00000176973), score: -0.37 FAM8A1family with sequence similarity 8, member A1 (ENSG00000137414), score: -0.4 FANCAFanconi anemia, complementation group A (ENSG00000187741), score: -0.39 FASLGFas ligand (TNF superfamily, member 6) (ENSG00000117560), score: 0.39 FBXO10F-box protein 10 (ENSG00000147912), score: 0.38 FBXO6F-box protein 6 (ENSG00000116663), score: -0.44 FBXW2F-box and WD repeat domain containing 2 (ENSG00000119402), score: -0.52 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: 0.38 FLGfilaggrin (ENSG00000143631), score: 0.49 FOXH1forkhead box H1 (ENSG00000160973), score: 0.52 FRMD4AFERM domain containing 4A (ENSG00000151474), score: 0.36 GABPB2GA binding protein transcription factor, beta subunit 2 (ENSG00000143458), score: 0.48 GBP3guanylate binding protein 3 (ENSG00000117226), score: 0.36 GCET2germinal center expressed transcript 2 (ENSG00000174500), score: 0.53 GDF9growth differentiation factor 9 (ENSG00000164404), score: -0.39 GINS4GINS complex subunit 4 (Sld5 homolog) (ENSG00000147536), score: 0.36 GIT2G protein-coupled receptor kinase interacting ArfGAP 2 (ENSG00000139436), score: -0.39 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: -0.38 GLG1golgi glycoprotein 1 (ENSG00000090863), score: 0.43 GLTPglycolipid transfer protein (ENSG00000139433), score: -0.38 GMPPAGDP-mannose pyrophosphorylase A (ENSG00000144591), score: 0.42 GNGT2guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 (ENSG00000167083), score: 0.46 GORABgolgin, RAB6-interacting (ENSG00000120370), score: 0.36 GORASP1golgi reassembly stacking protein 1, 65kDa (ENSG00000114745), score: 0.37 GP1BAglycoprotein Ib (platelet), alpha polypeptide (ENSG00000185245), score: 0.35 GPATCH1G patch domain containing 1 (ENSG00000076650), score: 0.36 GPR19G protein-coupled receptor 19 (ENSG00000183150), score: 0.63 GPX7glutathione peroxidase 7 (ENSG00000116157), score: 0.44 GSDMBgasdermin B (ENSG00000073605), score: 0.43 GTF2H3general transcription factor IIH, polypeptide 3, 34kDa (ENSG00000111358), score: 0.39 GTF2Igeneral transcription factor IIi (ENSG00000077809), score: -0.42 GTPBP5GTP binding protein 5 (putative) (ENSG00000101181), score: -0.44 GUCA1Bguanylate cyclase activator 1B (retina) (ENSG00000112599), score: -0.4 GUF1GUF1 GTPase homolog (S. cerevisiae) (ENSG00000151806), score: 0.5 GZMHgranzyme H (cathepsin G-like 2, protein h-CCPX) (ENSG00000100450), score: 0.52 HARS2histidyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000112855), score: -0.44 HAS3hyaluronan synthase 3 (ENSG00000103044), score: -0.43 HAUS7HAUS augmin-like complex, subunit 7 (ENSG00000213397), score: 0.38 HAUS8HAUS augmin-like complex, subunit 8 (ENSG00000131351), score: 0.39 HBE1hemoglobin, epsilon 1 (ENSG00000213931), score: 0.47 HCLS1hematopoietic cell-specific Lyn substrate 1 (ENSG00000180353), score: 0.38 HCRTR1hypocretin (orexin) receptor 1 (ENSG00000121764), score: -0.43 HDDC2HD domain containing 2 (ENSG00000111906), score: 0.43 HESX1HESX homeobox 1 (ENSG00000163666), score: 0.38 HHLA1HERV-H LTR-associating 1 (ENSG00000132297), score: 0.48 HIST1H1Chistone cluster 1, H1c (ENSG00000187837), score: 0.47 HIST1H2BJhistone cluster 1, H2bj (ENSG00000124635), score: 0.37 HIST2H2AChistone cluster 2, H2ac (ENSG00000184260), score: -0.37 HKDC1hexokinase domain containing 1 (ENSG00000156510), score: 0.83 HM13histocompatibility (minor) 13 (ENSG00000101294), score: -0.38 HMHA1histocompatibility (minor) HA-1 (ENSG00000180448), score: 0.38 HSD17B4hydroxysteroid (17-beta) dehydrogenase 4 (ENSG00000133835), score: -0.94 HSPA13heat shock protein 70kDa family, member 13 (ENSG00000155304), score: -0.44 HUWE1HECT, UBA and WWE domain containing 1 (ENSG00000086758), score: -0.82 HYIhydroxypyruvate isomerase (putative) (ENSG00000178922), score: 0.48 ICAM3intercellular adhesion molecule 3 (ENSG00000076662), score: 0.37 IFI44Linterferon-induced protein 44-like (ENSG00000137959), score: 0.51 IFNB1interferon, beta 1, fibroblast (ENSG00000171855), score: 0.38 IL17RAinterleukin 17 receptor A (ENSG00000177663), score: 0.5 IL4I1interleukin 4 induced 1 (ENSG00000104951), score: 0.42 INCA1inhibitor of CDK, cyclin A1 interacting protein 1 (ENSG00000196388), score: 0.47 INPP5Kinositol polyphosphate-5-phosphatase K (ENSG00000132376), score: -0.39 IP6K3inositol hexakisphosphate kinase 3 (ENSG00000161896), score: 0.75 ISCUiron-sulfur cluster scaffold homolog (E. coli) (ENSG00000136003), score: 0.36 ISXintestine-specific homeobox (ENSG00000175329), score: 0.42 ITGA11integrin, alpha 11 (ENSG00000137809), score: 0.77 JKAMPJNK1/MAPK8-associated membrane protein (ENSG00000050130), score: -0.39 KCNK7potassium channel, subfamily K, member 7 (ENSG00000173338), score: 0.58 KCTD20potassium channel tetramerisation domain containing 20 (ENSG00000112078), score: -0.46 KIAA0467KIAA0467 (ENSG00000198198), score: 0.55 KIF15kinesin family member 15 (ENSG00000163808), score: 0.6 KIF20Akinesin family member 20A (ENSG00000112984), score: 0.38 KIF3Bkinesin family member 3B (ENSG00000101350), score: -0.36 KINKIN, antigenic determinant of recA protein homolog (mouse) (ENSG00000151657), score: 0.36 KLHDC10kelch domain containing 10 (ENSG00000128607), score: -0.48 KLHL1kelch-like 1 (Drosophila) (ENSG00000150361), score: 0.99 KLRK1killer cell lectin-like receptor subfamily K, member 1 (ENSG00000213809), score: 0.42 KPNA6karyopherin alpha 6 (importin alpha 7) (ENSG00000025800), score: -0.44 LAIR2leukocyte-associated immunoglobulin-like receptor 2 (ENSG00000167618), score: 0.57 LASS6LAG1 homolog, ceramide synthase 6 (ENSG00000172292), score: -0.37 LBRlamin B receptor (ENSG00000143815), score: 0.36 LHFPL2lipoma HMGIC fusion partner-like 2 (ENSG00000145685), score: 0.37 LILRA4leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 (ENSG00000239961), score: 0.57 LINGO4leucine rich repeat and Ig domain containing 4 (ENSG00000213171), score: 0.53 LOC100130633hypothetical protein LOC100130633 (ENSG00000163867), score: -0.44 LOC100289938similar to cytoplasmic tRNA 2-thiolation protein 2 (ENSG00000174177), score: 0.53 LOC728129putative LRRC3-like protein ENSP00000367157 (ENSG00000204913), score: 0.51 LPXNleupaxin (ENSG00000110031), score: 0.39 LRCH3leucine-rich repeats and calponin homology (CH) domain containing 3 (ENSG00000186001), score: 0.48 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (ENSG00000198799), score: 0.4 LRIT3leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 (ENSG00000183423), score: 0.39 LRPAP1low density lipoprotein receptor-related protein associated protein 1 (ENSG00000163956), score: 0.52 LRRC40leucine rich repeat containing 40 (ENSG00000066557), score: -0.38 LRRC61leucine rich repeat containing 61 (ENSG00000127399), score: 0.36 LUC7L2LUC7-like 2 (S. cerevisiae) (ENSG00000146963), score: 0.48 LY75lymphocyte antigen 75 (ENSG00000054219), score: 0.73 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: 0.42 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: 0.71 MANSC1MANSC domain containing 1 (ENSG00000111261), score: -0.38 MAP2K4mitogen-activated protein kinase kinase 4 (ENSG00000065559), score: -0.37 MASTLmicrotubule associated serine/threonine kinase-like (ENSG00000120539), score: -0.39 MBD4methyl-CpG binding domain protein 4 (ENSG00000129071), score: 0.39 MBOAT4membrane bound O-acyltransferase domain containing 4 (ENSG00000177669), score: 0.43 MCOLN2mucolipin 2 (ENSG00000153898), score: 0.4 MED16mediator complex subunit 16 (ENSG00000175221), score: -0.42 MED9mediator complex subunit 9 (ENSG00000141026), score: 0.5 MEGF6multiple EGF-like-domains 6 (ENSG00000162591), score: 0.44 METTL10methyltransferase like 10 (ENSG00000203791), score: -0.59 MFSD2Bmajor facilitator superfamily domain containing 2B (ENSG00000205639), score: 0.58 MKNK1MAP kinase interacting serine/threonine kinase 1 (ENSG00000079277), score: 0.37 MOBKL3MOB1, Mps One Binder kinase activator-like 3 (yeast) (ENSG00000115540), score: 0.48 MORN4MORN repeat containing 4 (ENSG00000171160), score: 0.48 MPV17MpV17 mitochondrial inner membrane protein (ENSG00000115204), score: 0.43 MPV17LMPV17 mitochondrial membrane protein-like (ENSG00000156968), score: -0.47 MRPS17mitochondrial ribosomal protein S17 (ENSG00000239789), score: -0.72 MRPS23mitochondrial ribosomal protein S23 (ENSG00000181610), score: 0.4 MT1Fmetallothionein 1F (ENSG00000198417), score: -0.4 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (ENSG00000117983), score: 0.46 MXRA7matrix-remodelling associated 7 (ENSG00000182534), score: 0.41 MYCBPAPMYCBP associated protein (ENSG00000136449), score: 0.36 MYL6Bmyosin, light chain 6B, alkali, smooth muscle and non-muscle (ENSG00000196465), score: 0.62 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.42 NAALADL1N-acetylated alpha-linked acidic dipeptidase-like 1 (ENSG00000168060), score: 0.41 NAT15N-acetyltransferase 15 (GCN5-related, putative) (ENSG00000122390), score: -0.78 NAT9N-acetyltransferase 9 (GCN5-related, putative) (ENSG00000109065), score: -0.6 NDFIP1Nedd4 family interacting protein 1 (ENSG00000131507), score: -0.39 NDUFA9NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa (ENSG00000139180), score: -0.37 NDUFB2NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa (ENSG00000090266), score: -0.75 NEK6NIMA (never in mitosis gene a)-related kinase 6 (ENSG00000119408), score: 0.39 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: -0.52 NIPA2non imprinted in Prader-Willi/Angelman syndrome 2 (ENSG00000140157), score: -0.38 NKX2-6NK2 transcription factor related, locus 6 (Drosophila) (ENSG00000180053), score: 0.53 NLRP1NLR family, pyrin domain containing 1 (ENSG00000091592), score: 0.71 NME7non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000143156), score: -0.62 NMNAT1nicotinamide nucleotide adenylyltransferase 1 (ENSG00000173614), score: 0.35 NMT1N-myristoyltransferase 1 (ENSG00000136448), score: -0.37 NOL6nucleolar protein family 6 (RNA-associated) (ENSG00000165271), score: 0.36 NOL7nucleolar protein 7, 27kDa (ENSG00000225921), score: -0.38 NRBP2nuclear receptor binding protein 2 (ENSG00000185189), score: 0.43 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: 0.43 OPLAH5-oxoprolinase (ATP-hydrolysing) (ENSG00000178814), score: -0.36 OPN1SWopsin 1 (cone pigments), short-wave-sensitive (ENSG00000128617), score: -0.37 OR10G2olfactory receptor, family 10, subfamily G, member 2 (ENSG00000169202), score: 0.37 OR52N4olfactory receptor, family 52, subfamily N, member 4 (ENSG00000181074), score: 0.46 OSGEPL1O-sialoglycoprotein endopeptidase-like 1 (ENSG00000128694), score: 0.41 OSMoncostatin M (ENSG00000099985), score: 0.62 OXR1oxidation resistance 1 (ENSG00000164830), score: 0.37 PAN3PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (ENSG00000152520), score: 0.36 PARGpoly (ADP-ribose) glycohydrolase (ENSG00000227345), score: -0.44 PARP3poly (ADP-ribose) polymerase family, member 3 (ENSG00000041880), score: -0.39 PCID2PCI domain containing 2 (ENSG00000126226), score: 0.57 PDAP1PDGFA associated protein 1 (ENSG00000106244), score: 0.48 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.54 PDSS1prenyl (decaprenyl) diphosphate synthase, subunit 1 (ENSG00000148459), score: -0.74 PHF10PHD finger protein 10 (ENSG00000130024), score: -0.55 PITPNC1phosphatidylinositol transfer protein, cytoplasmic 1 (ENSG00000154217), score: 0.38 PIWIL3piwi-like 3 (Drosophila) (ENSG00000184571), score: 0.97 PKNOX1PBX/knotted 1 homeobox 1 (ENSG00000160199), score: -0.39 PLA2R1phospholipase A2 receptor 1, 180kDa (ENSG00000153246), score: 0.43 PLEKHH2pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 (ENSG00000152527), score: 0.39 PNKPpolynucleotide kinase 3'-phosphatase (ENSG00000039650), score: 0.37 POLMpolymerase (DNA directed), mu (ENSG00000122678), score: 0.37 POLR2Fpolymerase (RNA) II (DNA directed) polypeptide F (ENSG00000100142), score: 0.39 POLR3Epolymerase (RNA) III (DNA directed) polypeptide E (80kD) (ENSG00000058600), score: -0.51 PPIL3peptidylprolyl isomerase (cyclophilin)-like 3 (ENSG00000240344), score: -0.43 PPP1R16Aprotein phosphatase 1, regulatory (inhibitor) subunit 16A (ENSG00000160972), score: 0.45 PPPDE2PPPDE peptidase domain containing 2 (ENSG00000100418), score: -0.46 PRELPproline/arginine-rich end leucine-rich repeat protein (ENSG00000188783), score: 0.38 PRR4proline rich 4 (lacrimal) (ENSG00000111215), score: 0.71 PRXperiaxin (ENSG00000105227), score: -0.45 PSMA3proteasome (prosome, macropain) subunit, alpha type, 3 (ENSG00000100567), score: -0.4 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (ENSG00000159377), score: 0.4 PSMB7proteasome (prosome, macropain) subunit, beta type, 7 (ENSG00000136930), score: 0.46 PTCD1pentatricopeptide repeat domain 1 (ENSG00000106246), score: 0.82 PTPLBprotein tyrosine phosphatase-like (proline instead of catalytic arginine), member b (ENSG00000206527), score: -0.41 PYHIN1pyrin and HIN domain family, member 1 (ENSG00000163564), score: 0.43 PYROXD1pyridine nucleotide-disulphide oxidoreductase domain 1 (ENSG00000121350), score: -0.4 QRICH2glutamine rich 2 (ENSG00000129646), score: 0.45 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.36 RAB4ARAB4A, member RAS oncogene family (ENSG00000168118), score: -0.42 RAD9ARAD9 homolog A (S. pombe) (ENSG00000172613), score: 0.39 RALAv-ral simian leukemia viral oncogene homolog A (ras related) (ENSG00000006451), score: -0.85 RBM14RNA binding motif protein 14 (ENSG00000239306), score: -0.45 RBX1ring-box 1 (ENSG00000100387), score: -0.51 RCCD1RCC1 domain containing 1 (ENSG00000166965), score: -0.47 RCN3reticulocalbin 3, EF-hand calcium binding domain (ENSG00000142552), score: 0.46 RCOR1REST corepressor 1 (ENSG00000089902), score: -0.58 RCVRNrecoverin (ENSG00000109047), score: 0.46 RDM1RAD52 motif 1 (ENSG00000187456), score: 0.39 RECQL4RecQ protein-like 4 (ENSG00000160957), score: 0.58 REP15RAB15 effector protein (ENSG00000174236), score: 0.37 RNF103ring finger protein 103 (ENSG00000239305), score: 0.47 RNF160ring finger protein 160 (ENSG00000198862), score: -0.43 RNF26ring finger protein 26 (ENSG00000173456), score: 0.4 RP1L1retinitis pigmentosa 1-like 1 (ENSG00000183638), score: 0.42 RPF2ribosome production factor 2 homolog (S. cerevisiae) (ENSG00000197498), score: -0.62 RQCD1RCD1 required for cell differentiation1 homolog (S. pombe) (ENSG00000144580), score: -0.43 RRHretinal pigment epithelium-derived rhodopsin homolog (ENSG00000180245), score: 0.52 RSF1remodeling and spacing factor 1 (ENSG00000048649), score: -0.43 RUFY4RUN and FYVE domain containing 4 (ENSG00000188282), score: 0.38 SAMD11sterile alpha motif domain containing 11 (ENSG00000187634), score: 0.4 SAP18Sin3A-associated protein, 18kDa (ENSG00000150459), score: -0.41 SCNM1sodium channel modifier 1 (ENSG00000163156), score: 0.42 SELLselectin L (ENSG00000188404), score: 0.38 SERINC3serine incorporator 3 (ENSG00000132824), score: -0.48 SIRT3sirtuin 3 (ENSG00000142082), score: 0.4 SLC23A2solute carrier family 23 (nucleobase transporters), member 2 (ENSG00000089057), score: -0.45 SLC25A34solute carrier family 25, member 34 (ENSG00000162461), score: 0.5 SLC27A3solute carrier family 27 (fatty acid transporter), member 3 (ENSG00000143554), score: 0.39 SLC35B3solute carrier family 35, member B3 (ENSG00000124786), score: 0.36 SLURP1secreted LY6/PLAUR domain containing 1 (ENSG00000126233), score: 0.37 SNRNP27small nuclear ribonucleoprotein 27kDa (U4/U6.U5) (ENSG00000124380), score: -0.8 SNX18sorting nexin 18 (ENSG00000178996), score: -0.89 SNX33sorting nexin 33 (ENSG00000173548), score: 0.59 SPDYCspeedy homolog C (Xenopus laevis) (ENSG00000204710), score: 0.62 SPINK4serine peptidase inhibitor, Kazal type 4 (ENSG00000122711), score: 0.63 SPNsialophorin (ENSG00000197471), score: 0.93 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: 0.43 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.48 SSSCA1Sjogren syndrome/scleroderma autoantigen 1 (ENSG00000173465), score: 0.47 ST8SIA6ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 (ENSG00000148488), score: 0.49 STAMBPSTAM binding protein (ENSG00000124356), score: 0.37 STRAPserine/threonine kinase receptor associated protein (ENSG00000023734), score: -0.4 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.36 SUDS3suppressor of defective silencing 3 homolog (S. cerevisiae) (ENSG00000111707), score: -1 SULT1C4sulfotransferase family, cytosolic, 1C, member 4 (ENSG00000198075), score: 0.49 SVIPsmall VCP/p97-interacting protein (ENSG00000198168), score: -0.61 SYNCsyncoilin, intermediate filament protein (ENSG00000162520), score: -0.44 SYNCRIPsynaptotagmin binding, cytoplasmic RNA interacting protein (ENSG00000135316), score: -0.5 TACR2tachykinin receptor 2 (ENSG00000075073), score: 0.37 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (ENSG00000172660), score: 0.47 TAF1CTATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa (ENSG00000103168), score: 0.39 TAS1R3taste receptor, type 1, member 3 (ENSG00000169962), score: 0.5 TAS2R20taste receptor, type 2, member 20 (ENSG00000212125), score: 0.49 TAS2R3taste receptor, type 2, member 3 (ENSG00000127362), score: 0.39 TAS2R40taste receptor, type 2, member 40 (ENSG00000221937), score: 0.45 TAS2R5taste receptor, type 2, member 5 (ENSG00000127366), score: 0.61 TATDN2TatD DNase domain containing 2 (ENSG00000157014), score: 0.36 TBC1D20TBC1 domain family, member 20 (ENSG00000125875), score: -0.53 TBX10T-box 10 (ENSG00000167800), score: 0.54 TCTAT-cell leukemia translocation altered gene (ENSG00000145022), score: 0.38 TCTEX1D4Tctex1 domain containing 4 (ENSG00000188396), score: 0.38 THADAthyroid adenoma associated (ENSG00000115970), score: -0.41 THNSL2threonine synthase-like 2 (S. cerevisiae) (ENSG00000144115), score: 0.41 THSD1thrombospondin, type I, domain containing 1 (ENSG00000136114), score: 0.42 THTPAthiamine triphosphatase (ENSG00000157306), score: -0.5 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.36 TIMM17Atranslocase of inner mitochondrial membrane 17 homolog A (yeast) (ENSG00000134375), score: -0.4 TIPRLTIP41, TOR signaling pathway regulator-like (S. cerevisiae) (ENSG00000143155), score: -0.69 TM2D1TM2 domain containing 1 (ENSG00000162604), score: 0.39 TM2D3TM2 domain containing 3 (ENSG00000184277), score: -0.36 TM7SF4transmembrane 7 superfamily member 4 (ENSG00000164935), score: 0.41 TMCO6transmembrane and coiled-coil domains 6 (ENSG00000113119), score: 0.51 TMED6transmembrane emp24 protein transport domain containing 6 (ENSG00000157315), score: 0.37 TMEM104transmembrane protein 104 (ENSG00000109066), score: -0.37 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: -0.39 TMEM189transmembrane protein 189 (ENSG00000124208), score: -0.68 TMEM199transmembrane protein 199 (ENSG00000244045), score: 0.4 TMEM41Btransmembrane protein 41B (ENSG00000166471), score: -0.48 TMEM42transmembrane protein 42 (ENSG00000169964), score: 0.44 TMEM54transmembrane protein 54 (ENSG00000121900), score: 0.36 TMEM92transmembrane protein 92 (ENSG00000167105), score: 0.41 TMEM98transmembrane protein 98 (ENSG00000006042), score: 0.36 TMEM9BTMEM9 domain family, member B (ENSG00000175348), score: 0.45 TMPRSS9transmembrane protease, serine 9 (ENSG00000178297), score: -0.45 TNFAIP1tumor necrosis factor, alpha-induced protein 1 (endothelial) (ENSG00000109079), score: 0.7 TNFRSF17tumor necrosis factor receptor superfamily, member 17 (ENSG00000048462), score: 0.97 TNMDtenomodulin (ENSG00000000005), score: 0.48 TNNC2troponin C type 2 (fast) (ENSG00000101470), score: 0.39 TNPO1transportin 1 (ENSG00000083312), score: -0.94 TPM3tropomyosin 3 (ENSG00000143549), score: 0.36 TRAPPC10trafficking protein particle complex 10 (ENSG00000160218), score: -0.43 TRAPPC6Btrafficking protein particle complex 6B (ENSG00000182400), score: -0.36 TRIB2tribbles homolog 2 (Drosophila) (ENSG00000071575), score: 0.36 TRIM44tripartite motif-containing 44 (ENSG00000166326), score: 0.43 TRMT61BtRNA methyltransferase 61 homolog B (S. cerevisiae) (ENSG00000171103), score: -0.51 TSC22D1TSC22 domain family, member 1 (ENSG00000102804), score: -0.87 TSEN2tRNA splicing endonuclease 2 homolog (S. cerevisiae) (ENSG00000154743), score: 0.41 TSHRthyroid stimulating hormone receptor (ENSG00000165409), score: 0.65 TSPAN10tetraspanin 10 (ENSG00000182612), score: 0.36 TTC33tetratricopeptide repeat domain 33 (ENSG00000113638), score: -0.58 TTI1Tel2 interacting protein 1 homolog (S. pombe) (ENSG00000101407), score: 0.37 TTLtubulin tyrosine ligase (ENSG00000114999), score: -0.43 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (ENSG00000117143), score: -0.86 UBBubiquitin B (ENSG00000170315), score: 0.4 UBE2CBPubiquitin-conjugating enzyme E2C binding protein (ENSG00000118420), score: 0.59 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: 0.36 UBE3Bubiquitin protein ligase E3B (ENSG00000151148), score: 0.42 UBL3ubiquitin-like 3 (ENSG00000122042), score: -0.4 UBR7ubiquitin protein ligase E3 component n-recognin 7 (putative) (ENSG00000012963), score: -0.49 UBXN8UBX domain protein 8 (ENSG00000104691), score: -0.4 UMODL1uromodulin-like 1 (ENSG00000177398), score: 0.38 USP12ubiquitin specific peptidase 12 (ENSG00000152484), score: -0.64 USP9Xubiquitin specific peptidase 9, X-linked (ENSG00000124486), score: -0.49 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.61 VPS16vacuolar protein sorting 16 homolog (S. cerevisiae) (ENSG00000215305), score: -0.38 VSIG10V-set and immunoglobulin domain containing 10 (ENSG00000176834), score: 0.4 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: -0.48 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (ENSG00000196584), score: 0.74 XRCC3X-ray repair complementing defective repair in Chinese hamster cells 3 (ENSG00000126215), score: 0.4 YJEFN3YjeF N-terminal domain containing 3 (ENSG00000186010), score: 0.41 YPEL5yippee-like 5 (Drosophila) (ENSG00000119801), score: -0.53 ZCWPW1zinc finger, CW type with PWWP domain 1 (ENSG00000078487), score: -0.53 ZDHHC3zinc finger, DHHC-type containing 3 (ENSG00000163812), score: -0.43 ZFYVE1zinc finger, FYVE domain containing 1 (ENSG00000165861), score: -0.37 ZFYVE21zinc finger, FYVE domain containing 21 (ENSG00000100711), score: 0.45 ZMAT5zinc finger, matrin type 5 (ENSG00000100319), score: 0.38 ZNF169zinc finger protein 169 (ENSG00000175787), score: 0.41 ZNF180zinc finger protein 180 (ENSG00000167384), score: 0.36 ZNF211zinc finger protein 211 (ENSG00000121417), score: -0.48 ZNF24zinc finger protein 24 (ENSG00000172466), score: 0.62 ZNF267zinc finger protein 267 (ENSG00000185947), score: 0.45 ZNF275zinc finger protein 275 (ENSG00000063587), score: -0.48 ZNF323zinc finger protein 323 (ENSG00000235109), score: 0.4 ZNF385Czinc finger protein 385C (ENSG00000187595), score: -0.36 ZNF500zinc finger protein 500 (ENSG00000103199), score: -0.45 ZNF514zinc finger protein 514 (ENSG00000144026), score: -0.51 ZNF557zinc finger protein 557 (ENSG00000130544), score: 0.39 ZNF575zinc finger protein 575 (ENSG00000176472), score: 0.39 ZNF592zinc finger protein 592 (ENSG00000166716), score: -0.47 ZNF615zinc finger protein 615 (ENSG00000197619), score: 0.64 ZNF641zinc finger protein 641 (ENSG00000167528), score: 0.37 ZNF7zinc finger protein 7 (ENSG00000147789), score: 0.41 ZNF707zinc finger protein 707 (ENSG00000181135), score: 0.39 ZNF783zinc finger family member 783 (ENSG00000204946), score: 0.37 ZNF789zinc finger protein 789 (ENSG00000198556), score: 0.42 ZNF83zinc finger protein 83 (ENSG00000167766), score: -0.4 ZSCAN1zinc finger and SCAN domain containing 1 (ENSG00000152467), score: 0.36 ZSWIM7zinc finger, SWIM-type containing 7 (ENSG00000214941), score: 0.38 ZWINTZW10 interactor (ENSG00000122952), score: -0.46
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_ts_m_ca1 | ggo | ts | m | _ |
| ggo_br_f_ca1 | ggo | br | f | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ggo_br_m_ca1 | ggo | br | m | _ |
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| ggo_kd_m_ca1 | ggo | kd | m | _ |
| ggo_kd_f_ca1 | ggo | kd | f | _ |
| ggo_lv_f_ca1 | ggo | lv | f | _ |
| ggo_lv_m_ca1 | ggo | lv | m | _ |
| ggo_ht_f_ca1 | ggo | ht | f | _ |
| ggo_ht_m_ca1 | ggo | ht | m | _ |