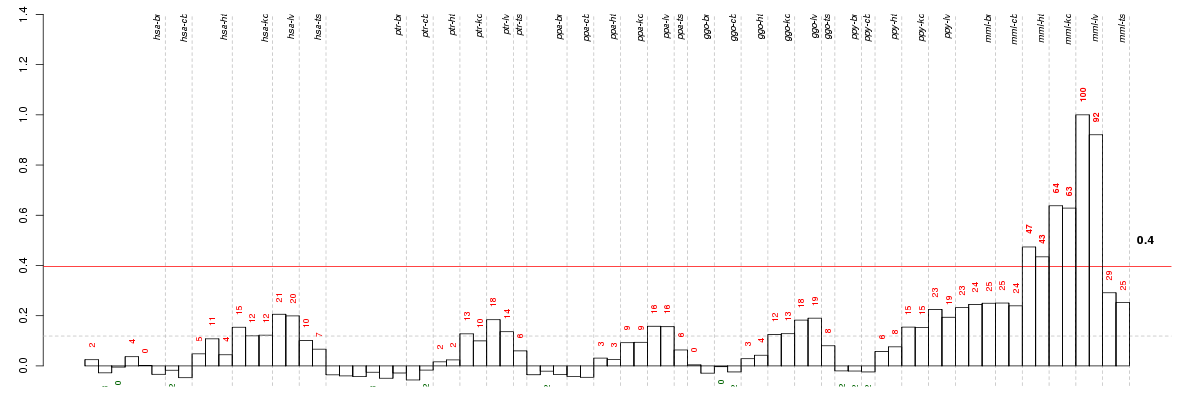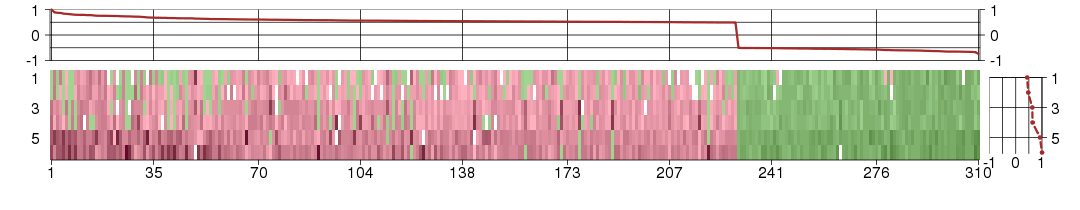

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
membrane lipid metabolic process
The chemical reactions and pathways involving membrane lipids, any lipid found in or associated with a biological membrane.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
membrane lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of membrane lipids, any lipid found in or associated with a biological membrane.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, as carried out by individual cells.
cellular lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, as carried out by individual cells.
lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
membrane lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of membrane lipids, any lipid found in or associated with a biological membrane.


A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.62 ABCA4ATP-binding cassette, sub-family A (ABC1), member 4 (ENSG00000198691), score: 0.68 ABP1amiloride binding protein 1 (amine oxidase (copper-containing)) (ENSG00000002726), score: 0.55 ABTB2ankyrin repeat and BTB (POZ) domain containing 2 (ENSG00000166016), score: 0.55 ACCS1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) (ENSG00000110455), score: 0.53 ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.55 ACOX1acyl-CoA oxidase 1, palmitoyl (ENSG00000161533), score: 0.53 ADAM12ADAM metallopeptidase domain 12 (ENSG00000148848), score: 0.87 ADIPOR1adiponectin receptor 1 (ENSG00000159346), score: 0.52 AFPalpha-fetoprotein (ENSG00000081051), score: 0.8 AGPHD1aminoglycoside phosphotransferase domain containing 1 (ENSG00000188266), score: 0.57 AGR3anterior gradient homolog 3 (Xenopus laevis) (ENSG00000173467), score: 0.52 AIM1Labsent in melanoma 1-like (ENSG00000176092), score: 0.63 AIM2absent in melanoma 2 (ENSG00000163568), score: 0.54 AIREautoimmune regulator (ENSG00000160224), score: 0.54 AKR1CL1aldo-keto reductase family 1, member C-like 1 (ENSG00000196326), score: 0.79 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: 0.67 ALOX12arachidonate 12-lipoxygenase (ENSG00000108839), score: 0.79 ANKDD1Aankyrin repeat and death domain containing 1A (ENSG00000166839), score: -0.56 ANO7anoctamin 7 (ENSG00000146205), score: 0.6 ANXA10annexin A10 (ENSG00000109511), score: 0.54 AP3M2adaptor-related protein complex 3, mu 2 subunit (ENSG00000070718), score: -0.54 APOA4apolipoprotein A-IV (ENSG00000110244), score: 0.77 APRTadenine phosphoribosyltransferase (ENSG00000198931), score: 0.5 AQP8aquaporin 8 (ENSG00000103375), score: 0.53 ARAP1ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 (ENSG00000186635), score: 0.54 ARCN1archain 1 (ENSG00000095139), score: 0.56 ARSBarylsulfatase B (ENSG00000113273), score: 0.56 ART4ADP-ribosyltransferase 4 (Dombrock blood group) (ENSG00000111339), score: 0.62 AS3MTarsenic (+3 oxidation state) methyltransferase (ENSG00000214435), score: 0.52 ATP8ATP synthase F0 subunit 8 (ENSG00000228253), score: -0.61 ATPBD4ATP binding domain 4 (ENSG00000134146), score: 0.52 B3GAT2beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) (ENSG00000112309), score: -0.65 B3GNT7UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 (ENSG00000156966), score: 0.68 BCAR3breast cancer anti-estrogen resistance 3 (ENSG00000137936), score: 0.58 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (ENSG00000083123), score: 0.54 BIKBCL2-interacting killer (apoptosis-inducing) (ENSG00000100290), score: 0.51 BIRC7baculoviral IAP repeat-containing 7 (ENSG00000101197), score: 0.74 BLMHbleomycin hydrolase (ENSG00000108578), score: 0.52 C14orf126chromosome 14 open reading frame 126 (ENSG00000129480), score: 0.56 C15orf57chromosome 15 open reading frame 57 (ENSG00000128891), score: -0.62 C16orf45chromosome 16 open reading frame 45 (ENSG00000166780), score: -0.56 C16orf91chromosome 16 open reading frame 91 (ENSG00000174109), score: 0.68 C17orf49chromosome 17 open reading frame 49 (ENSG00000161939), score: 0.55 C1orf177chromosome 1 open reading frame 177 (ENSG00000162398), score: 0.62 C20orf29chromosome 20 open reading frame 29 (ENSG00000125843), score: 0.6 C20orf7chromosome 20 open reading frame 7 (ENSG00000101247), score: -0.66 C22orf15chromosome 22 open reading frame 15 (ENSG00000169314), score: 0.73 C22orf39chromosome 22 open reading frame 39 (ENSG00000242259), score: -0.55 C2orf43chromosome 2 open reading frame 43 (ENSG00000118961), score: 0.55 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.57 C3orf26chromosome 3 open reading frame 26 (ENSG00000184220), score: -0.59 C3orf37chromosome 3 open reading frame 37 (ENSG00000183624), score: -0.57 C5orf4chromosome 5 open reading frame 4 (ENSG00000170271), score: 0.53 C6orf108chromosome 6 open reading frame 108 (ENSG00000112667), score: -0.56 C6orf52chromosome 6 open reading frame 52 (ENSG00000137434), score: 0.6 C9orf152chromosome 9 open reading frame 152 (ENSG00000188959), score: 0.79 CABP7calcium binding protein 7 (ENSG00000100314), score: -0.53 CAMTA1calmodulin binding transcription activator 1 (ENSG00000171735), score: -0.65 CASP10caspase 10, apoptosis-related cysteine peptidase (ENSG00000003400), score: 0.5 CASP9caspase 9, apoptosis-related cysteine peptidase (ENSG00000132906), score: 0.53 CBARA1calcium binding atopy-related autoantigen 1 (ENSG00000107745), score: -0.51 CBY1chibby homolog 1 (Drosophila) (ENSG00000100211), score: -0.54 CCDC115coiled-coil domain containing 115 (ENSG00000136710), score: 0.54 CCDC142coiled-coil domain containing 142 (ENSG00000135637), score: 0.65 CCDC28Bcoiled-coil domain containing 28B (ENSG00000160050), score: -0.54 CCDC88Ccoiled-coil domain containing 88C (ENSG00000015133), score: 0.49 CCL23chemokine (C-C motif) ligand 23 (ENSG00000167236), score: 0.72 CCL24chemokine (C-C motif) ligand 24 (ENSG00000106178), score: 0.66 CCL25chemokine (C-C motif) ligand 25 (ENSG00000131142), score: 0.61 CCR3chemokine (C-C motif) receptor 3 (ENSG00000183625), score: 0.56 CD1DCD1d molecule (ENSG00000158473), score: 0.51 CD1ECD1e molecule (ENSG00000158488), score: 0.78 CD3GCD3g molecule, gamma (CD3-TCR complex) (ENSG00000160654), score: 0.5 CDC42BPACDC42 binding protein kinase alpha (DMPK-like) (ENSG00000143776), score: -0.67 CDIPTCDP-diacylglycerol--inositol 3-phosphatidyltransferase (ENSG00000103502), score: 0.52 CDK5cyclin-dependent kinase 5 (ENSG00000164885), score: -0.56 CDKN2AIPNLCDKN2A interacting protein N-terminal like (ENSG00000237190), score: -0.61 CDV3CDV3 homolog (mouse) (ENSG00000091527), score: -0.63 CDX2caudal type homeobox 2 (ENSG00000165556), score: 0.58 CHCHD2coiled-coil-helix-coiled-coil-helix domain containing 2 (ENSG00000106153), score: -0.58 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.61 CLDN14claudin 14 (ENSG00000159261), score: 0.49 CLDN15claudin 15 (ENSG00000106404), score: 0.56 CLINT1clathrin interactor 1 (ENSG00000113282), score: 0.61 CNIHcornichon homolog (Drosophila) (ENSG00000100528), score: 0.5 CREG1cellular repressor of E1A-stimulated genes 1 (ENSG00000143162), score: 0.51 CROTcarnitine O-octanoyltransferase (ENSG00000005469), score: 0.5 CRYBA2crystallin, beta A2 (ENSG00000163499), score: 0.75 CXCR2chemokine (C-X-C motif) receptor 2 (ENSG00000180871), score: 0.6 CXCR3chemokine (C-X-C motif) receptor 3 (ENSG00000186810), score: 0.66 CYFIP1cytoplasmic FMR1 interacting protein 1 (ENSG00000068793), score: 0.59 CYP1A1cytochrome P450, family 1, subfamily A, polypeptide 1 (ENSG00000140465), score: 0.6 DEFB4Bdefensin, beta 4B (ENSG00000171711), score: 0.52 DEGS1degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) (ENSG00000143753), score: 0.53 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: -0.57 DISP1dispatched homolog 1 (Drosophila) (ENSG00000154309), score: 0.54 DKK2dickkopf homolog 2 (Xenopus laevis) (ENSG00000155011), score: 0.7 DLK1delta-like 1 homolog (Drosophila) (ENSG00000185559), score: 0.53 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (ENSG00000168259), score: -0.62 DNASE2Bdeoxyribonuclease II beta (ENSG00000137976), score: 0.54 DPYDdihydropyrimidine dehydrogenase (ENSG00000188641), score: 0.54 DSC3desmocollin 3 (ENSG00000134762), score: 0.74 DTX4deltex homolog 4 (Drosophila) (ENSG00000110042), score: -0.55 EARS2glutamyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000103356), score: 0.53 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (ENSG00000134109), score: 0.5 ELMOD3ELMO/CED-12 domain containing 3 (ENSG00000115459), score: 0.61 EME1essential meiotic endonuclease 1 homolog 1 (S. pombe) (ENSG00000154920), score: 0.51 EMR1egf-like module containing, mucin-like, hormone receptor-like 1 (ENSG00000174837), score: 0.58 ERLIN2ER lipid raft associated 2 (ENSG00000147475), score: 0.58 ESPL1extra spindle pole bodies homolog 1 (S. cerevisiae) (ENSG00000135476), score: 0.5 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.87 FAM120Afamily with sequence similarity 120A (ENSG00000048828), score: 0.51 FAM120AOSfamily with sequence similarity 120A opposite strand (ENSG00000188938), score: 0.61 FAM189Bfamily with sequence similarity 189, member B (ENSG00000160767), score: -0.74 FGD2FYVE, RhoGEF and PH domain containing 2 (ENSG00000146192), score: 0.52 FKTNfukutin (ENSG00000106692), score: 0.52 FLJ35220hypothetical protein FLJ35220 (ENSG00000173818), score: -0.63 FOXP3forkhead box P3 (ENSG00000049768), score: 0.66 FPR3formyl peptide receptor 3 (ENSG00000187474), score: 0.57 FXC1fracture callus 1 homolog (rat) (ENSG00000132286), score: 0.62 FXYD6FXYD domain containing ion transport regulator 6 (ENSG00000137726), score: -0.52 FZD5frizzled homolog 5 (Drosophila) (ENSG00000163251), score: 0.51 GAS8growth arrest-specific 8 (ENSG00000141013), score: 0.51 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.59 GGT5gamma-glutamyltransferase 5 (ENSG00000099998), score: -0.52 GLYATL3glycine-N-acyltransferase-like 3 (ENSG00000203972), score: 0.65 GNAT1guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 (ENSG00000114349), score: 0.63 GNAT2guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 (ENSG00000134183), score: 0.56 GPC6glypican 6 (ENSG00000183098), score: 0.52 GPR113G protein-coupled receptor 113 (ENSG00000173567), score: 0.54 GPR152G protein-coupled receptor 152 (ENSG00000175514), score: 0.54 GPR172BG protein-coupled receptor 172B (ENSG00000132517), score: 0.59 GSTA4glutathione S-transferase alpha 4 (ENSG00000170899), score: -0.51 GTF2F1general transcription factor IIF, polypeptide 1, 74kDa (ENSG00000125651), score: -0.65 GTSE1G-2 and S-phase expressed 1 (ENSG00000075218), score: 0.53 GYLTL1Bglycosyltransferase-like 1B (ENSG00000165905), score: 0.57 HAUS3HAUS augmin-like complex, subunit 3 (ENSG00000214367), score: -0.51 HEATR3HEAT repeat containing 3 (ENSG00000155393), score: 0.58 HEATR4HEAT repeat containing 4 (ENSG00000187105), score: -0.51 HFEhemochromatosis (ENSG00000010704), score: 0.57 HNRNPLheterogeneous nuclear ribonucleoprotein L (ENSG00000104824), score: -0.66 HRASLS2HRAS-like suppressor 2 (ENSG00000133328), score: 0.83 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (ENSG00000153976), score: 0.53 HSDL1hydroxysteroid dehydrogenase like 1 (ENSG00000103160), score: -0.54 IFIT1Binterferon-induced protein with tetratricopeptide repeats 1B (ENSG00000204010), score: 0.75 IGFL3IGF-like family member 3 (ENSG00000188624), score: 0.56 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.5 IL2interleukin 2 (ENSG00000109471), score: 0.49 INHBAinhibin, beta A (ENSG00000122641), score: 0.52 INO80EINO80 complex subunit E (ENSG00000169592), score: -0.6 ITFG2integrin alpha FG-GAP repeat containing 2 (ENSG00000111203), score: 0.53 IVDisovaleryl-CoA dehydrogenase (ENSG00000128928), score: 0.61 JMJD4jumonji domain containing 4 (ENSG00000081692), score: 0.55 KAZALD1Kazal-type serine peptidase inhibitor domain 1 (ENSG00000107821), score: 0.49 KBTBD2kelch repeat and BTB (POZ) domain containing 2 (ENSG00000170852), score: -0.52 KIAA0141KIAA0141 (ENSG00000081791), score: 0.52 KIAA1609KIAA1609 (ENSG00000140950), score: 0.55 KIAA1841KIAA1841 (ENSG00000162929), score: -0.6 KIAA1967KIAA1967 (ENSG00000158941), score: -0.52 KIFAP3kinesin-associated protein 3 (ENSG00000075945), score: -0.51 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.66 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.53 KRT28keratin 28 (ENSG00000173908), score: 0.84 LGMNlegumain (ENSG00000100600), score: 0.59 LIPMlipase, family member M (ENSG00000173239), score: 0.61 LMBR1limb region 1 homolog (mouse) (ENSG00000105983), score: -0.6 LOC100127905family with sequence similarity 165, member B pseudogene (ENSG00000198738), score: 0.75 LRIG3leucine-rich repeats and immunoglobulin-like domains 3 (ENSG00000139263), score: 0.49 LRRC8Eleucine rich repeat containing 8 family, member E (ENSG00000171017), score: 0.52 LUC7LLUC7-like (S. cerevisiae) (ENSG00000007392), score: -0.63 LYPD3LY6/PLAUR domain containing 3 (ENSG00000124466), score: 0.67 MAGEF1melanoma antigen family F, 1 (ENSG00000177383), score: -0.54 MAKmale germ cell-associated kinase (ENSG00000111837), score: 0.52 MAP3K12mitogen-activated protein kinase kinase kinase 12 (ENSG00000139625), score: -0.53 MAP3K7mitogen-activated protein kinase kinase kinase 7 (ENSG00000135341), score: -0.53 MAT2Bmethionine adenosyltransferase II, beta (ENSG00000038274), score: 0.49 MCFD2multiple coagulation factor deficiency 2 (ENSG00000180398), score: 0.51 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.72 MEFVMediterranean fever (ENSG00000103313), score: 0.56 MPV17LMPV17 mitochondrial membrane protein-like (ENSG00000156968), score: 0.58 MRPS14mitochondrial ribosomal protein S14 (ENSG00000120333), score: 0.5 MS4A14membrane-spanning 4-domains, subfamily A, member 14 (ENSG00000166928), score: 0.52 MS4A8Bmembrane-spanning 4-domains, subfamily A, member 8B (ENSG00000166959), score: 0.64 MSLNmesothelin (ENSG00000102854), score: 0.59 MSLNLmesothelin-like (ENSG00000162006), score: 0.6 NAA40N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) (ENSG00000110583), score: -0.6 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.49 NET1neuroepithelial cell transforming 1 (ENSG00000173848), score: 0.53 NGDNneuroguidin, EIF4E binding protein (ENSG00000129460), score: -0.51 NGRNneugrin, neurite outgrowth associated (ENSG00000182768), score: -0.65 NIPAL1NIPA-like domain containing 1 (ENSG00000163293), score: 0.53 NME6non-metastatic cells 6, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000172113), score: 0.58 NSDHLNAD(P) dependent steroid dehydrogenase-like (ENSG00000147383), score: 0.57 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.62 NUDT15nudix (nucleoside diphosphate linked moiety X)-type motif 15 (ENSG00000136159), score: -0.59 NUDT21nudix (nucleoside diphosphate linked moiety X)-type motif 21 (ENSG00000167005), score: -0.55 NUMBnumb homolog (Drosophila) (ENSG00000133961), score: 0.55 NUPL2nucleoporin like 2 (ENSG00000136243), score: -0.51 NYXnyctalopin (ENSG00000188937), score: 0.55 OLA1Obg-like ATPase 1 (ENSG00000138430), score: -0.52 OPLAH5-oxoprolinase (ATP-hydrolysing) (ENSG00000178814), score: 0.52 OPTNoptineurin (ENSG00000123240), score: -0.51 OR10H3olfactory receptor, family 10, subfamily H, member 3 (ENSG00000171936), score: 0.63 ORC4Lorigin recognition complex, subunit 4-like (S. cerevisiae) (ENSG00000115947), score: -0.54 OTUD1OTU domain containing 1 (ENSG00000165312), score: 0.55 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.61 P2RY2purinergic receptor P2Y, G-protein coupled, 2 (ENSG00000175591), score: 0.54 PAEPprogestagen-associated endometrial protein (ENSG00000122133), score: 0.69 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.56 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (ENSG00000102981), score: -0.54 PARP15poly (ADP-ribose) polymerase family, member 15 (ENSG00000173200), score: 0.68 PARS2prolyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000162396), score: 0.49 PDCD2programmed cell death 2 (ENSG00000071994), score: 0.52 PLA2G12Aphospholipase A2, group XIIA (ENSG00000123739), score: 0.53 PLA2G2Dphospholipase A2, group IID (ENSG00000117215), score: 0.89 PLEKHB2pleckstrin homology domain containing, family B (evectins) member 2 (ENSG00000115762), score: -0.65 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.64 PMPCBpeptidase (mitochondrial processing) beta (ENSG00000105819), score: 0.5 POLR3Epolymerase (RNA) III (DNA directed) polypeptide E (80kD) (ENSG00000058600), score: 0.49 PPAPDC1Bphosphatidic acid phosphatase type 2 domain containing 1B (ENSG00000147535), score: 0.62 PRDM4PR domain containing 4 (ENSG00000110851), score: -0.55 PRHOXNBparahox cluster neighbor (ENSG00000183463), score: 0.82 PROL1proline rich, lacrimal 1 (ENSG00000171199), score: 0.57 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.79 PSKH2protein serine kinase H2 (ENSG00000147613), score: 0.75 PTCD2pentatricopeptide repeat domain 2 (ENSG00000049883), score: 0.52 QRSL1glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 (ENSG00000130348), score: 0.51 RAB1BRAB1B, member RAS oncogene family (ENSG00000174903), score: -0.65 RANBP10RAN binding protein 10 (ENSG00000141084), score: 0.55 RBM6RNA binding motif protein 6 (ENSG00000004534), score: -0.6 RCBTB2regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 (ENSG00000136161), score: 0.51 REC8REC8 homolog (yeast) (ENSG00000100918), score: -0.52 REPS1RALBP1 associated Eps domain containing 1 (ENSG00000135597), score: -0.52 RGS12regulator of G-protein signaling 12 (ENSG00000159788), score: -0.54 RGS13regulator of G-protein signaling 13 (ENSG00000127074), score: 0.73 RHBDD3rhomboid domain containing 3 (ENSG00000100263), score: 0.56 RNASE1ribonuclease, RNase A family, 1 (pancreatic) (ENSG00000129538), score: -0.6 RNF149ring finger protein 149 (ENSG00000163162), score: 0.5 RPL36ALribosomal protein L36a-like (ENSG00000165502), score: -0.64 RPN1ribophorin I (ENSG00000163902), score: 0.49 RPP40ribonuclease P/MRP 40kDa subunit (ENSG00000124787), score: 0.58 SATL1spermidine/spermine N1-acetyl transferase-like 1 (ENSG00000184788), score: -0.56 SCLT1sodium channel and clathrin linker 1 (ENSG00000151466), score: 0.71 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.63 SEC24ASEC24 family, member A (S. cerevisiae) (ENSG00000113615), score: 0.5 SEC31ASEC31 homolog A (S. cerevisiae) (ENSG00000138674), score: 0.51 SEPT12septin 12 (ENSG00000140623), score: 0.51 SERINC4serine incorporator 4 (ENSG00000184716), score: 0.78 SERPINA12serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 (ENSG00000165953), score: 0.66 SIP1survival of motor neuron protein interacting protein 1 (ENSG00000092208), score: -0.61 SLC15A5solute carrier family 15, member 5 (ENSG00000188991), score: 0.54 SLC16A11solute carrier family 16, member 11 (monocarboxylic acid transporter 11) (ENSG00000174326), score: 0.59 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.64 SLC25A14solute carrier family 25 (mitochondrial carrier, brain), member 14 (ENSG00000102078), score: -0.56 SLC45A3solute carrier family 45, member 3 (ENSG00000158715), score: 0.53 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.56 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.54 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (ENSG00000166311), score: 0.51 SMPDL3Asphingomyelin phosphodiesterase, acid-like 3A (ENSG00000172594), score: 0.51 SNUPNsnurportin 1 (ENSG00000169371), score: -0.51 SOAT2sterol O-acyltransferase 2 (ENSG00000167780), score: 0.56 SRD5A3steroid 5 alpha-reductase 3 (ENSG00000128039), score: 0.65 SRP14signal recognition particle 14kDa (homologous Alu RNA binding protein) (ENSG00000140319), score: -0.66 SRSF6serine/arginine-rich splicing factor 6 (ENSG00000124193), score: 0.56 STARD5StAR-related lipid transfer (START) domain containing 5 (ENSG00000172345), score: 0.51 SULT1C4sulfotransferase family, cytosolic, 1C, member 4 (ENSG00000198075), score: 0.58 TBX19T-box 19 (ENSG00000143178), score: 0.75 TCEA2transcription elongation factor A (SII), 2 (ENSG00000171703), score: -0.66 TCOF1Treacher Collins-Franceschetti syndrome 1 (ENSG00000070814), score: -0.53 TEP1telomerase-associated protein 1 (ENSG00000129566), score: 0.52 TLR6toll-like receptor 6 (ENSG00000174130), score: 0.75 TMEM104transmembrane protein 104 (ENSG00000109066), score: 0.52 TMEM179Btransmembrane protein 179B (ENSG00000185475), score: 0.53 TMEM19transmembrane protein 19 (ENSG00000139291), score: 0.54 TMEM30Btransmembrane protein 30B (ENSG00000182107), score: 0.51 TMEM55Atransmembrane protein 55A (ENSG00000155099), score: -0.51 TMEM61transmembrane protein 61 (ENSG00000143001), score: 0.56 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.51 TNFSF15tumor necrosis factor (ligand) superfamily, member 15 (ENSG00000181634), score: 0.53 TNNtenascin N (ENSG00000120332), score: 0.62 TRPM5transient receptor potential cation channel, subfamily M, member 5 (ENSG00000070985), score: 0.73 TSKUtsukushi small leucine rich proteoglycan homolog (Xenopus laevis) (ENSG00000182704), score: 0.5 TTC21Btetratricopeptide repeat domain 21B (ENSG00000123607), score: -0.58 TTC24tetratricopeptide repeat domain 24 (ENSG00000187862), score: 0.54 TTC9Ctetratricopeptide repeat domain 9C (ENSG00000162222), score: -0.53 UBXN11UBX domain protein 11 (ENSG00000158062), score: -0.53 UCMAupper zone of growth plate and cartilage matrix associated (ENSG00000165623), score: 0.57 UCN3urocortin 3 (stresscopin) (ENSG00000178473), score: 0.52 VWCEvon Willebrand factor C and EGF domains (ENSG00000167992), score: 0.55 WDR55WD repeat domain 55 (ENSG00000120314), score: 0.56 XPCxeroderma pigmentosum, complementation group C (ENSG00000154767), score: 0.58 ZBED2zinc finger, BED-type containing 2 (ENSG00000177494), score: 0.55 ZBED3zinc finger, BED-type containing 3 (ENSG00000132846), score: 0.55 ZBTB3zinc finger and BTB domain containing 3 (ENSG00000185670), score: 0.54 ZMYND19zinc finger, MYND-type containing 19 (ENSG00000165724), score: -0.51 ZNF114zinc finger protein 114 (ENSG00000178150), score: 0.61 ZNF146zinc finger protein 146 (ENSG00000167635), score: -0.61 ZNF16zinc finger protein 16 (ENSG00000170631), score: 0.58 ZNF226zinc finger protein 226 (ENSG00000167380), score: -0.56 ZNF496zinc finger protein 496 (ENSG00000162714), score: -0.57 ZNF618zinc finger protein 618 (ENSG00000157657), score: 0.5 ZNHIT3zinc finger, HIT type 3 (ENSG00000108278), score: -0.61 ZSWIM3zinc finger, SWIM-type containing 3 (ENSG00000132801), score: 0.59
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_ht_f_ca1 | mml | ht | f | _ |
| mml_ht_m_ca1 | mml | ht | m | _ |
| mml_kd_f_ca1 | mml | kd | f | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |