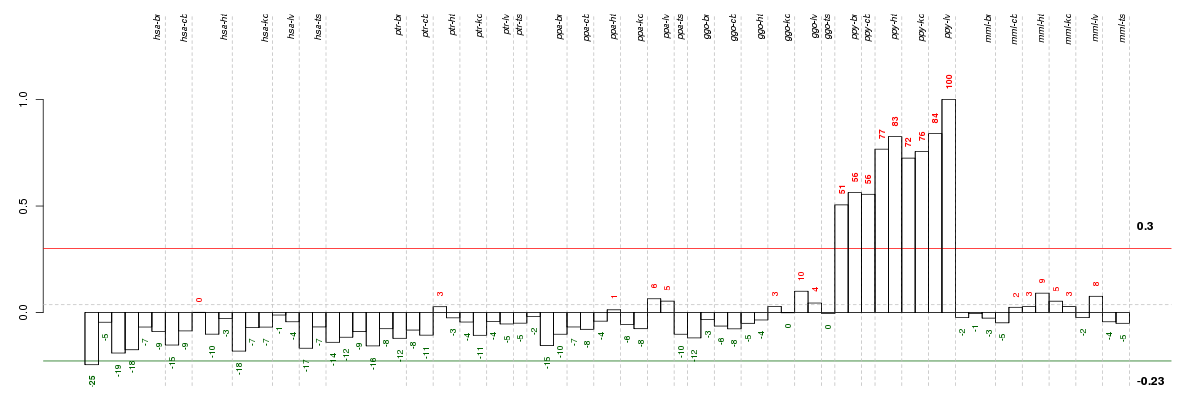

Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
cell killing
Any process in an organism that results in the killing of its own cells or those of another organism, including in some cases the death of the other organism. Killing here refers to the induction of death in one cell by another cell, not cell-autonomous death due to internal or other environmental conditions.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II
The process by which an antigen-presenting cell expresses antigen (peptide or polysaccharide) on its cell surface in association with an MHC class II protein complex.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
RNA processing
Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules.
translation
The cellular metabolic process by which a protein is formed, using the sequence of a mature mRNA molecule to specify the sequence of amino acids in a polypeptide chain. Translation is mediated by the ribosome, and begins with the formation of a ternary complex between aminoacylated initiator methionine tRNA, GTP, and initiation factor 2, which subsequently associates with the small subunit of the ribosome and an mRNA. Translation ends with the release of a polypeptide chain from the ribosome.
translational initiation
The process preceding formation of the peptide bond between the first two amino acids of a protein. This includes the formation of a complex of the ribosome, mRNA, and an initiation complex that contains the first aminoacyl-tRNA.
regulation of translation
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA.
regulation of translational initiation
Any process that modulates the frequency, rate or extent of translational initiation.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
viral reproduction
The process by which a virus reproduces. Usually, this is by infection of a host cell, replication of the viral genome, and assembly of progeny virus particles. In some cases the viral genetic material may integrate into the host genome and only subsequently, under particular circumstances, 'complete' its life cycle.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
posttranscriptional regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression after the production of an RNA transcript.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
virus-host interaction
Interactions, directly with the host cell macromolecular machinery, to allow virus replication.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
antigen processing and presentation
The process by which an antigen-presenting cell expresses antigen (peptide or lipid) on its cell surface in association with an MHC protein complex.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
viral reproductive process
A reproductive process involved in viral reproduction. Usually, this is by infection of a host cell, replication of the viral genome, and assembly of progeny virus particles. In some cases the viral genetic material may integrate into the host genome and only subsequently, under particular circumstances, 'complete' its life cycle.
symbiosis, encompassing mutualism through parasitism
An interaction between two organisms living together in more or less intimate association. The term host is usually used for the larger (macro) of the two members of a symbiosis. The smaller (micro) member is called the symbiont organism. Microscopic symbionts are often referred to as endosymbionts. The various forms of symbiosis include parasitism, in which the association is disadvantageous or destructive to one of the organisms; mutualism, in which the association is advantageous, or often necessary to one or both and not harmful to either; and commensalism, in which one member of the association benefits while the other is not affected. However, mutualism, parasitism, and commensalism are often not discrete categories of interactions and should rather be perceived as a continuum of interaction ranging from parasitism to mutualism. In fact, the direction of a symbiotic interaction can change during the lifetime of the symbionts due to developmental changes as well as changes in the biotic/abiotic environment in which the interaction occurs.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
killing of cells of another organism
Any process in an organism that results in the killing of cells of another organism, including in some cases the death of the other organism. Killing here refers to the induction of death in one cell by another cell, not cell-autonomous death due to internal or other environmental conditions.
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of translation in response to stress
Modulation of the frequency, rate or extent of translation as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
regulation of translational initiation in response to stress
Any process that activates or increases the frequency, rate or extent of translation initiation, as a result of a stimulus indicating the organism is under stress.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
interspecies interaction between organisms
Any process by which an organism has an effect on an organism of a different species.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein.
interaction with host
An interaction between two organisms living together in more or less intimate association. The term host is used for the larger (macro) of the two members of a symbiosis; the various forms of symbiosis include parasitism, commensalism and mutualism.
interaction with symbiont
An interaction between two organisms living together in more or less intimate association. The term symbiont is used for the smaller (macro) of the two members of a symbiosis; the various forms of symbiosis include parasitism, commensalism and mutualism.
multi-organism process
Any process by which an organism has an effect on another organism of the same or different species.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
modification of morphology or physiology of other organism involved in symbiotic interaction
The process by which an organism effects a change in the structure or processes of a second organism, where the two organisms are in a symbiotic interaction.
disruption of cells of other organism involved in symbiotic interaction
A process by which an organism has a negative effect on the functioning of the second organism's cells, where the two organisms are in a symbiotic interaction.
modification by host of symbiont morphology or physiology
The process by which an organism effects a change in the structure or processes of a symbiont organism. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
disruption by host of symbiont cells
Any process by which an organism has a negative effect on the functioning of the symbiont's cells. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
killing by host of symbiont cells
Any process mediated by an organism that results in the death of cells in the symbiont organism. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
killing of cells in other organism involved in symbiotic interaction
Any process mediated by an organism that results in the death of cells in a second organism, where the two organisms are in a symbiotic interaction.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
viral reproductive process
A reproductive process involved in viral reproduction. Usually, this is by infection of a host cell, replication of the viral genome, and assembly of progeny virus particles. In some cases the viral genetic material may integrate into the host genome and only subsequently, under particular circumstances, 'complete' its life cycle.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
killing of cells of another organism
Any process in an organism that results in the killing of cells of another organism, including in some cases the death of the other organism. Killing here refers to the induction of death in one cell by another cell, not cell-autonomous death due to internal or other environmental conditions.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
interaction with host
An interaction between two organisms living together in more or less intimate association. The term host is used for the larger (macro) of the two members of a symbiosis; the various forms of symbiosis include parasitism, commensalism and mutualism.
interaction with symbiont
An interaction between two organisms living together in more or less intimate association. The term symbiont is used for the smaller (macro) of the two members of a symbiosis; the various forms of symbiosis include parasitism, commensalism and mutualism.
modification of morphology or physiology of other organism involved in symbiotic interaction
The process by which an organism effects a change in the structure or processes of a second organism, where the two organisms are in a symbiotic interaction.
virus-host interaction
Interactions, directly with the host cell macromolecular machinery, to allow virus replication.
modification by host of symbiont morphology or physiology
The process by which an organism effects a change in the structure or processes of a symbiont organism. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
translation
The cellular metabolic process by which a protein is formed, using the sequence of a mature mRNA molecule to specify the sequence of amino acids in a polypeptide chain. Translation is mediated by the ribosome, and begins with the formation of a ternary complex between aminoacylated initiator methionine tRNA, GTP, and initiation factor 2, which subsequently associates with the small subunit of the ribosome and an mRNA. Translation ends with the release of a polypeptide chain from the ribosome.
regulation of translation
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA.
regulation of translation
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA.
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
translational initiation
The process preceding formation of the peptide bond between the first two amino acids of a protein. This includes the formation of a complex of the ribosome, mRNA, and an initiation complex that contains the first aminoacyl-tRNA.
regulation of translation
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA.
translation
The cellular metabolic process by which a protein is formed, using the sequence of a mature mRNA molecule to specify the sequence of amino acids in a polypeptide chain. Translation is mediated by the ribosome, and begins with the formation of a ternary complex between aminoacylated initiator methionine tRNA, GTP, and initiation factor 2, which subsequently associates with the small subunit of the ribosome and an mRNA. Translation ends with the release of a polypeptide chain from the ribosome.
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
RNA processing
Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of translational initiation in response to stress
Any process that activates or increases the frequency, rate or extent of translation initiation, as a result of a stimulus indicating the organism is under stress.
disruption by host of symbiont cells
Any process by which an organism has a negative effect on the functioning of the symbiont's cells. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
killing of cells in other organism involved in symbiotic interaction
Any process mediated by an organism that results in the death of cells in a second organism, where the two organisms are in a symbiotic interaction.
regulation of translational initiation
Any process that modulates the frequency, rate or extent of translational initiation.
regulation of translation in response to stress
Modulation of the frequency, rate or extent of translation as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
regulation of translation
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA.
killing by host of symbiont cells
Any process mediated by an organism that results in the death of cells in the symbiont organism. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
lytic vacuole
A vacuole that is maintained at an acidic pH and which contains degradative enzymes, including a wide variety of acid hydrolases.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
MHC class II protein complex
A transmembrane protein complex composed of an MHC class II alpha and MHC class II beta chain, and with or without a bound peptide or polysaccharide antigen.
lysosome
A small lytic vacuole that has cell cycle-independent morphology and is found in most animal cells and that contains a variety of hydrolases, most of which have their maximal activities in the pH range 5-6. The contained enzymes display latency if properly isolated. About 40 different lysosomal hydrolases are known and lysosomes have a great variety of morphologies and functions.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
lysosomal membrane
The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm.
vacuolar membrane
The lipid bilayer surrounding the vacuole and separating its contents from the cytoplasm of the cell.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
eukaryotic translation initiation factor 2B complex
A multisubunit guanine nucleotide exchange factor which catalyzes the exchange of GDP bound to initiation factor eIF2 for GTP, generating active eIF2-GTP. In humans, it is composed of five subunits, alpha, beta, delta, gamma and epsilon.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
integral to endoplasmic reticulum membrane
Penetrating at least one phospholipid bilayer of an endoplasmic reticulum membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
organelle membrane
The lipid bilayer surrounding an organelle.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to endoplasmic reticulum membrane
Located in the endoplasmic reticulum membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to organelle membrane
Penetrating at least one phospholipid bilayer of an organelle membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
MHC protein complex
A transmembrane protein complex composed of an MHC alpha chain and, in most cases, either an MHC class II beta chain or an invariant beta2-microglobin chain, and with or without a bound peptide, lipid, or polysaccharide antigen.
MHC class I peptide loading complex
A large, multisubunit complex which consists of the MHC class I-beta 2 microglobulin dimer, the transporter associated with antigen presentation (TAP), tapasin (an MHC-encoded membrane protein), the chaperone calreticulin and the thiol oxidoreductase ERp57. Functions in the assembly of peptides with newly synthesized MHC class I molecules.
TAP complex
A heterodimer composed of the subunits TAP1 and TAP2 (transporter associated with antigen presentation). Functions in the transport of antigenic peptides from the cytosol to the lumen of the endoplasmic reticulum.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
vacuolar part
Any constituent part of a vacuole, a closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
nuclear membrane-endoplasmic reticulum network
The continuous network of membranes encompassing the outer nuclear membrane and the endoplasmic reticulum.
TAP complex
A heterodimer composed of the subunits TAP1 and TAP2 (transporter associated with antigen presentation). Functions in the transport of antigenic peptides from the cytosol to the lumen of the endoplasmic reticulum.
intrinsic to endoplasmic reticulum membrane
Located in the endoplasmic reticulum membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
intrinsic to endoplasmic reticulum membrane
Located in the endoplasmic reticulum membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
MHC class I peptide loading complex
A large, multisubunit complex which consists of the MHC class I-beta 2 microglobulin dimer, the transporter associated with antigen presentation (TAP), tapasin (an MHC-encoded membrane protein), the chaperone calreticulin and the thiol oxidoreductase ERp57. Functions in the assembly of peptides with newly synthesized MHC class I molecules.
TAP complex
A heterodimer composed of the subunits TAP1 and TAP2 (transporter associated with antigen presentation). Functions in the transport of antigenic peptides from the cytosol to the lumen of the endoplasmic reticulum.
vacuolar membrane
The lipid bilayer surrounding the vacuole and separating its contents from the cytoplasm of the cell.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
endoplasmic reticulum membrane
The lipid bilayer surrounding the endoplasmic reticulum.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
endoplasmic reticulum
The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached).
eukaryotic translation initiation factor 2B complex
A multisubunit guanine nucleotide exchange factor which catalyzes the exchange of GDP bound to initiation factor eIF2 for GTP, generating active eIF2-GTP. In humans, it is composed of five subunits, alpha, beta, delta, gamma and epsilon.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
vacuolar part
Any constituent part of a vacuole, a closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material.
subsynaptic reticulum
An elaborate tubulolamellar membrane system that underlies the postsynaptic cell membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
MHC protein complex
A transmembrane protein complex composed of an MHC alpha chain and, in most cases, either an MHC class II beta chain or an invariant beta2-microglobin chain, and with or without a bound peptide, lipid, or polysaccharide antigen.
vacuolar part
Any constituent part of a vacuole, a closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
endoplasmic reticulum part
Any constituent part of the endoplasmic reticulum, the irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae.
integral to endoplasmic reticulum membrane
Penetrating at least one phospholipid bilayer of an endoplasmic reticulum membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
integral to organelle membrane
Penetrating at least one phospholipid bilayer of an organelle membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
MHC class I peptide loading complex
A large, multisubunit complex which consists of the MHC class I-beta 2 microglobulin dimer, the transporter associated with antigen presentation (TAP), tapasin (an MHC-encoded membrane protein), the chaperone calreticulin and the thiol oxidoreductase ERp57. Functions in the assembly of peptides with newly synthesized MHC class I molecules.
lysosomal membrane
The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm.

protein binding
Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
RNA binding
Interacting selectively and non-covalently with an RNA molecule or a portion thereof.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
MHC class II receptor activity
Combining with an MHC class II protein complex to initiate a change in cellular activity.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
TAP binding
Interacting selectively and non-covalently with TAP protein, transporter associated with antigen processing protein. TAP protein is a heterodimeric peptide transporter consisting of the subunits TAP1 and TAP2.
TAP1 binding
Interacting selectively and non-covalently with the TAP1 subunit of TAP (transporter associated with antigen processing) protein.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05322 | 4.248e-13 | 1.735 | 19 | 52 | Systemic lupus erythematosus |
| 04612 | 7.365e-06 | 1.468 | 11 | 44 | Antigen processing and presentation |
| 05330 | 3.657e-04 | 0.8008 | 7 | 24 | Allograft rejection |
| 05332 | 3.657e-04 | 0.8008 | 7 | 24 | Graft-versus-host disease |
| 04940 | 7.501e-04 | 0.9009 | 7 | 27 | Type I diabetes mellitus |
| 05320 | 9.389e-04 | 0.9343 | 7 | 28 | Autoimmune thyroid disease |
| 05310 | 1.495e-03 | 0.7007 | 6 | 21 | Asthma |
| 04672 | 4.871e-03 | 1.235 | 7 | 37 | Intestinal immune network for IgA production |
| 05416 | 5.636e-03 | 1.268 | 7 | 38 | Viral myocarditis |
| 05140 | 2.606e-02 | 1.702 | 7 | 51 | Leishmaniasis |
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (ENSG00000064687), score: 0.46 ABHD12Babhydrolase domain containing 12B (ENSG00000131969), score: 0.61 ABI3BPABI family, member 3 (NESH) binding protein (ENSG00000154175), score: 0.48 ACCN4amiloride-sensitive cation channel 4, pituitary (ENSG00000072182), score: 0.6 ACOT13acyl-CoA thioesterase 13 (ENSG00000112304), score: 0.44 ACSBG1acyl-CoA synthetase bubblegum family member 1 (ENSG00000103740), score: -0.66 ADCK2aarF domain containing kinase 2 (ENSG00000133597), score: 0.94 AGERadvanced glycosylation end product-specific receptor (ENSG00000204305), score: 0.59 ALAS2aminolevulinate, delta-, synthase 2 (ENSG00000158578), score: 0.64 ALX3ALX homeobox 3 (ENSG00000156150), score: -0.49 ANGPT2angiopoietin 2 (ENSG00000091879), score: 0.44 ANKRD53ankyrin repeat domain 53 (ENSG00000144031), score: 0.49 API5apoptosis inhibitor 5 (ENSG00000166181), score: -0.94 ARF5ADP-ribosylation factor 5 (ENSG00000004059), score: -0.49 ARGLU1arginine and glutamate rich 1 (ENSG00000134884), score: 0.44 ARL1ADP-ribosylation factor-like 1 (ENSG00000120805), score: 0.56 ARPC4actin related protein 2/3 complex, subunit 4, 20kDa (ENSG00000214021), score: -0.47 ARR3arrestin 3, retinal (X-arrestin) (ENSG00000120500), score: 0.5 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.44 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.56 ASB14ankyrin repeat and SOCS box-containing 14 (ENSG00000239388), score: 0.55 ATRIPATR interacting protein (ENSG00000164053), score: 0.86 ATXN1ataxin 1 (ENSG00000124788), score: -0.63 AVILadvillin (ENSG00000135407), score: 0.79 AZU1azurocidin 1 (ENSG00000172232), score: 0.59 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (ENSG00000235863), score: -0.7 B3GAT2beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) (ENSG00000112309), score: 0.46 BATF3basic leucine zipper transcription factor, ATF-like 3 (ENSG00000123685), score: 0.45 BEST1bestrophin 1 (ENSG00000167995), score: -0.67 BIRC2baculoviral IAP repeat-containing 2 (ENSG00000110330), score: 0.49 BIRC3baculoviral IAP repeat-containing 3 (ENSG00000023445), score: 0.49 BPIL1bactericidal/permeability-increasing protein-like 1 (ENSG00000078898), score: 0.49 BRD2bromodomain containing 2 (ENSG00000204256), score: -1 BRD4bromodomain containing 4 (ENSG00000141867), score: -0.49 BRD7bromodomain containing 7 (ENSG00000166164), score: 0.44 BTLAB and T lymphocyte associated (ENSG00000186265), score: 0.44 C10orf2chromosome 10 open reading frame 2 (ENSG00000107815), score: -0.56 C10orf32chromosome 10 open reading frame 32 (ENSG00000166275), score: -0.48 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.44 C12orf45chromosome 12 open reading frame 45 (ENSG00000151131), score: 0.48 C12orf52chromosome 12 open reading frame 52 (ENSG00000139405), score: -0.53 C16orf48chromosome 16 open reading frame 48 (ENSG00000124074), score: -0.46 C16orf57chromosome 16 open reading frame 57 (ENSG00000103005), score: -0.57 C16orf61chromosome 16 open reading frame 61 (ENSG00000103121), score: 0.74 C16orf7chromosome 16 open reading frame 7 (ENSG00000075399), score: -0.59 C16orf79chromosome 16 open reading frame 79 (ENSG00000182685), score: -0.66 C17orf67chromosome 17 open reading frame 67 (ENSG00000214226), score: 0.51 C17orf76chromosome 17 open reading frame 76 (ENSG00000181350), score: -0.51 C17orf99chromosome 17 open reading frame 99 (ENSG00000187997), score: 0.45 C18orf22chromosome 18 open reading frame 22 (ENSG00000101546), score: -0.56 C1orf159chromosome 1 open reading frame 159 (ENSG00000131591), score: -0.5 C1orf192chromosome 1 open reading frame 192 (ENSG00000188931), score: -0.56 C1orf27chromosome 1 open reading frame 27 (ENSG00000157181), score: 0.56 C1orf31chromosome 1 open reading frame 31 (ENSG00000168275), score: 0.45 C1orf56chromosome 1 open reading frame 56 (ENSG00000143443), score: -0.6 C1QTNF5C1q and tumor necrosis factor related protein 5 (ENSG00000235718), score: 0.49 C20orf70chromosome 20 open reading frame 70 (ENSG00000131050), score: 0.48 C21orf2chromosome 21 open reading frame 2 (ENSG00000160226), score: -0.56 C2CD4BC2 calcium-dependent domain containing 4B (ENSG00000205502), score: 0.49 C2orf66chromosome 2 open reading frame 66 (ENSG00000187944), score: 0.66 C2orf79chromosome 2 open reading frame 79 (ENSG00000184924), score: -0.52 C3orf37chromosome 3 open reading frame 37 (ENSG00000183624), score: -0.51 C3orf75chromosome 3 open reading frame 75 (ENSG00000163832), score: -0.78 C4orf29chromosome 4 open reading frame 29 (ENSG00000164074), score: 0.52 C5orf13chromosome 5 open reading frame 13 (ENSG00000134986), score: -0.76 C6orf126chromosome 6 open reading frame 126 (ENSG00000196748), score: 0.7 C6orf162chromosome 6 open reading frame 162 (ENSG00000111850), score: 0.45 C7orf36chromosome 7 open reading frame 36 (ENSG00000241127), score: 0.46 C7orf53chromosome 7 open reading frame 53 (ENSG00000181016), score: 0.47 C9orf95chromosome 9 open reading frame 95 (ENSG00000106733), score: 0.52 CA1carbonic anhydrase I (ENSG00000133742), score: 0.69 CA5Bcarbonic anhydrase VB, mitochondrial (ENSG00000169239), score: 0.44 CABP7calcium binding protein 7 (ENSG00000100314), score: -0.49 CAMPcathelicidin antimicrobial peptide (ENSG00000164047), score: 0.49 CAPN12calpain 12 (ENSG00000182472), score: -0.49 CASP4caspase 4, apoptosis-related cysteine peptidase (ENSG00000196954), score: 0.48 CBX7chromobox homolog 7 (ENSG00000100307), score: -0.46 CBY1chibby homolog 1 (Drosophila) (ENSG00000100211), score: 0.44 CCDC152coiled-coil domain containing 152 (ENSG00000198865), score: -0.65 CCDC153coiled-coil domain containing 153 (ENSG00000204313), score: 0.45 CCDC42Bcoiled-coil domain containing 42B (ENSG00000186710), score: -0.58 CCDC77coiled-coil domain containing 77 (ENSG00000120647), score: -0.46 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.52 CCHCR1coiled-coil alpha-helical rod protein 1 (ENSG00000204536), score: -0.71 CCL7chemokine (C-C motif) ligand 7 (ENSG00000108688), score: 0.45 CCNG1cyclin G1 (ENSG00000113328), score: 0.45 CCT8chaperonin containing TCP1, subunit 8 (theta) (ENSG00000156261), score: 0.48 CD101CD101 molecule (ENSG00000134256), score: 0.62 CD209CD209 molecule (ENSG00000090659), score: 0.46 CD80CD80 molecule (ENSG00000121594), score: 0.52 CDC25Acell division cycle 25 homolog A (S. pombe) (ENSG00000164045), score: 0.48 CEBPECCAAT/enhancer binding protein (C/EBP), epsilon (ENSG00000092067), score: 0.7 CENPNcentromere protein N (ENSG00000166451), score: -0.64 CENPOcentromere protein O (ENSG00000138092), score: -0.64 CFDP1craniofacial development protein 1 (ENSG00000153774), score: 0.54 CHMP5chromatin modifying protein 5 (ENSG00000086065), score: 0.49 CHRNEcholinergic receptor, nicotinic, epsilon (ENSG00000108556), score: -0.52 CHSY1chondroitin sulfate synthase 1 (ENSG00000131873), score: 0.47 CIRBPcold inducible RNA binding protein (ENSG00000099622), score: -0.46 CKAP4cytoskeleton-associated protein 4 (ENSG00000136026), score: 0.51 CLEC5AC-type lectin domain family 5, member A (ENSG00000090269), score: 0.8 COL7A1collagen, type VII, alpha 1 (ENSG00000114270), score: -0.5 COMMD3COMM domain containing 3 (ENSG00000148444), score: 0.55 COMMD6COMM domain containing 6 (ENSG00000188243), score: 0.45 COQ3coenzyme Q3 homolog, methyltransferase (S. cerevisiae) (ENSG00000132423), score: 0.45 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.48 CTSGcathepsin G (ENSG00000100448), score: 0.44 CTSKcathepsin K (ENSG00000143387), score: 0.61 CYB561D1cytochrome b-561 domain containing 1 (ENSG00000174151), score: -0.55 CYP2U1cytochrome P450, family 2, subfamily U, polypeptide 1 (ENSG00000155016), score: 0.5 DAXXdeath-domain associated protein (ENSG00000204209), score: -0.96 DBF4BDBF4 homolog B (S. cerevisiae) (ENSG00000161692), score: -0.55 DCAF16DDB1 and CUL4 associated factor 16 (ENSG00000163257), score: -0.46 DCAF17DDB1 and CUL4 associated factor 17 (ENSG00000115827), score: -0.62 DCP1ADCP1 decapping enzyme homolog A (S. cerevisiae) (ENSG00000162290), score: -0.46 DDIT4DNA-damage-inducible transcript 4 (ENSG00000168209), score: 0.51 DDRGK1DDRGK domain containing 1 (ENSG00000198171), score: -0.48 DDX18DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 (ENSG00000088205), score: 0.55 DDX41DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (ENSG00000183258), score: -0.58 DDX5DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (ENSG00000108654), score: 0.49 DEM1defects in morphology 1 homolog (S. cerevisiae) (ENSG00000164002), score: 0.44 DIDO1death inducer-obliterator 1 (ENSG00000101191), score: 0.48 DNAH1dynein, axonemal, heavy chain 1 (ENSG00000114841), score: -0.5 DNAJC30DnaJ (Hsp40) homolog, subfamily C, member 30 (ENSG00000176410), score: -0.51 DNAL1dynein, axonemal, light chain 1 (ENSG00000119661), score: 0.7 DOC2Adouble C2-like domains, alpha (ENSG00000149927), score: -0.58 DONSONdownstream neighbor of SON (ENSG00000159147), score: -0.56 ECHDC1enoyl CoA hydratase domain containing 1 (ENSG00000093144), score: 0.55 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (ENSG00000172071), score: 0.51 EIF2B1eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa (ENSG00000111361), score: 0.44 EIF2B5eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa (ENSG00000145191), score: 0.63 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (ENSG00000134001), score: 0.5 ELANEelastase, neutrophil expressed (ENSG00000197561), score: 0.46 EME1essential meiotic endonuclease 1 homolog 1 (S. pombe) (ENSG00000154920), score: 0.47 EMG1EMG1 nucleolar protein homolog (S. cerevisiae) (ENSG00000126749), score: 0.45 EMR3egf-like module containing, mucin-like, hormone receptor-like 3 (ENSG00000131355), score: 0.79 ENY2enhancer of yellow 2 homolog (Drosophila) (ENSG00000120533), score: 0.45 ERCC5excision repair cross-complementing rodent repair deficiency, complementation group 5 (ENSG00000134899), score: 0.56 EXOSC3exosome component 3 (ENSG00000107371), score: 0.6 FAM103A1family with sequence similarity 103, member A1 (ENSG00000169612), score: 0.46 FAM134Cfamily with sequence similarity 134, member C (ENSG00000141699), score: -0.49 FAM162Afamily with sequence similarity 162, member A (ENSG00000114023), score: 0.53 FAM166Afamily with sequence similarity 166, member A (ENSG00000188163), score: -0.9 FAM173Bfamily with sequence similarity 173, member B (ENSG00000150756), score: 0.58 FAM174Bfamily with sequence similarity 174, member B (ENSG00000185442), score: -0.55 FAM185Afamily with sequence similarity 185, member A (ENSG00000222011), score: 0.44 FAM18B2family with sequence similarity 18, member B2 (ENSG00000239704), score: -0.52 FAM36Afamily with sequence similarity 36, member A (ENSG00000203667), score: 0.58 FBXO21F-box protein 21 (ENSG00000135108), score: -0.5 FGF2fibroblast growth factor 2 (basic) (ENSG00000138685), score: -0.74 FRMD8FERM domain containing 8 (ENSG00000126391), score: -0.77 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.47 GALNTL2UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2 (ENSG00000131386), score: -0.49 GCC2GRIP and coiled-coil domain containing 2 (ENSG00000135968), score: 0.49 GFM2G elongation factor, mitochondrial 2 (ENSG00000164347), score: 0.64 GHRHgrowth hormone releasing hormone (ENSG00000118702), score: 0.86 GINS2GINS complex subunit 2 (Psf2 homolog) (ENSG00000131153), score: -0.49 GLI4GLI family zinc finger 4 (ENSG00000181638), score: -0.53 GLIPR1GLI pathogenesis-related 1 (ENSG00000139278), score: 0.56 GLTPglycolipid transfer protein (ENSG00000139433), score: -0.69 GNGT1guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 (ENSG00000127928), score: 0.57 GPATCH4G patch domain containing 4 (ENSG00000160818), score: 0.45 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.65 GPR44G protein-coupled receptor 44 (ENSG00000183134), score: -0.53 GPR97G protein-coupled receptor 97 (ENSG00000182885), score: 0.47 GTF2E1general transcription factor IIE, polypeptide 1, alpha 56kDa (ENSG00000153767), score: 0.55 GTF3C1general transcription factor IIIC, polypeptide 1, alpha 220kDa (ENSG00000077235), score: -0.48 GZMKgranzyme K (granzyme 3; tryptase II) (ENSG00000113088), score: 0.5 HARS2histidyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000112855), score: 0.63 HAS3hyaluronan synthase 3 (ENSG00000103044), score: 0.46 HCCSholocytochrome c synthase (ENSG00000004961), score: 0.45 HIST1H1Bhistone cluster 1, H1b (ENSG00000184357), score: 0.52 HIST1H3Ahistone cluster 1, H3a (ENSG00000112727), score: 0.53 HLA-DMAmajor histocompatibility complex, class II, DM alpha (ENSG00000204257), score: -0.67 HLA-DMBmajor histocompatibility complex, class II, DM beta (ENSG00000242574), score: -0.63 HLA-DOBmajor histocompatibility complex, class II, DO beta (ENSG00000204267), score: -0.83 HLA-DPA1major histocompatibility complex, class II, DP alpha 1 (ENSG00000231389), score: -0.74 HLA-DPB1major histocompatibility complex, class II, DP beta 1 (ENSG00000223865), score: -0.77 HLA-DRAmajor histocompatibility complex, class II, DR alpha (ENSG00000204287), score: -0.78 HNRNPH3heterogeneous nuclear ribonucleoprotein H3 (2H9) (ENSG00000096746), score: 0.53 HPSEheparanase (ENSG00000173083), score: 0.45 HSD17B8hydroxysteroid (17-beta) dehydrogenase 8 (ENSG00000204228), score: -0.75 HSPA1Lheat shock 70kDa protein 1-like (ENSG00000204390), score: 0.46 HSPA5heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) (ENSG00000044574), score: 0.47 HTATIP2HIV-1 Tat interactive protein 2, 30kDa (ENSG00000109854), score: -0.6 HTThuntingtin (ENSG00000197386), score: -0.5 IBSPintegrin-binding sialoprotein (ENSG00000029559), score: 0.58 IGBP1immunoglobulin (CD79A) binding protein 1 (ENSG00000089289), score: 0.46 IL34interleukin 34 (ENSG00000157368), score: 0.47 IRF4interferon regulatory factor 4 (ENSG00000137265), score: 0.44 ITSN2intersectin 2 (ENSG00000198399), score: 0.46 JMJD6jumonji domain containing 6 (ENSG00000070495), score: 0.51 KIAA0195KIAA0195 (ENSG00000177728), score: -0.58 KIAA0892KIAA0892 (ENSG00000129933), score: -0.55 KRT27keratin 27 (ENSG00000171446), score: 0.54 LFNGLFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (ENSG00000106003), score: -0.5 LIFleukemia inhibitory factor (cholinergic differentiation factor) (ENSG00000128342), score: 0.49 LIG1ligase I, DNA, ATP-dependent (ENSG00000105486), score: -0.49 LIMD2LIM domain containing 2 (ENSG00000136490), score: -0.5 LIN7Clin-7 homolog C (C. elegans) (ENSG00000148943), score: 0.46 LONP2lon peptidase 2, peroxisomal (ENSG00000102910), score: 0.51 LOXL3lysyl oxidase-like 3 (ENSG00000115318), score: -0.78 LOXL4lysyl oxidase-like 4 (ENSG00000138131), score: 0.51 LRPAP1low density lipoprotein receptor-related protein associated protein 1 (ENSG00000163956), score: -0.48 LRRC23leucine rich repeat containing 23 (ENSG00000010626), score: -0.57 LRRC50leucine rich repeat containing 50 (ENSG00000154099), score: -0.46 LYG2lysozyme G-like 2 (ENSG00000185674), score: 0.83 MAGOHBmago-nashi homolog B (Drosophila) (ENSG00000111196), score: 0.52 MAPK12mitogen-activated protein kinase 12 (ENSG00000188130), score: -0.61 MAPKSP1MAPK scaffold protein 1 (ENSG00000109270), score: 0.45 MED17mediator complex subunit 17 (ENSG00000042429), score: 0.57 MED21mediator complex subunit 21 (ENSG00000152944), score: 0.45 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.53 MED6mediator complex subunit 6 (ENSG00000133997), score: -0.57 MEF2Bmyocyte enhancer factor 2B (ENSG00000064489), score: -0.7 MEMO1mediator of cell motility 1 (ENSG00000162961), score: 0.47 MESDC2mesoderm development candidate 2 (ENSG00000117899), score: 0.61 MFSD8major facilitator superfamily domain containing 8 (ENSG00000164073), score: 0.5 MINAMYC induced nuclear antigen (ENSG00000170854), score: 0.59 MIPOL1mirror-image polydactyly 1 (ENSG00000151338), score: 0.44 MITD1MIT, microtubule interacting and transport, domain containing 1 (ENSG00000158411), score: 0.56 MLANAmelan-A (ENSG00000120215), score: 0.68 MLST8MTOR associated protein, LST8 homolog (S. cerevisiae) (ENSG00000167965), score: -0.5 MMABmethylmalonic aciduria (cobalamin deficiency) cblB type (ENSG00000139428), score: 0.52 MMP8matrix metallopeptidase 8 (neutrophil collagenase) (ENSG00000118113), score: 0.49 MPHOSPH6M-phase phosphoprotein 6 (ENSG00000135698), score: 0.5 MRPL36mitochondrial ribosomal protein L36 (ENSG00000171421), score: -0.7 MRPL9mitochondrial ribosomal protein L9 (ENSG00000143436), score: 0.57 MRPS14mitochondrial ribosomal protein S14 (ENSG00000120333), score: 0.44 MRPS16mitochondrial ribosomal protein S16 (ENSG00000182180), score: 0.48 MRPS17mitochondrial ribosomal protein S17 (ENSG00000239789), score: 0.49 MRPS30mitochondrial ribosomal protein S30 (ENSG00000112996), score: -0.59 MRPS33mitochondrial ribosomal protein S33 (ENSG00000090263), score: 0.52 MSMBmicroseminoprotein, beta- (ENSG00000138294), score: 0.53 MTERFmitochondrial transcription termination factor (ENSG00000127989), score: 0.47 MTERFD2MTERF domain containing 2 (ENSG00000122085), score: 0.47 MTHFD2Lmethylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (ENSG00000163738), score: 0.5 MUS81MUS81 endonuclease homolog (S. cerevisiae) (ENSG00000172732), score: -0.49 MYOZ3myozenin 3 (ENSG00000164591), score: 0.62 MYSM1Myb-like, SWIRM and MPN domains 1 (ENSG00000162601), score: 0.53 N4BP2L2NEDD4 binding protein 2-like 2 (ENSG00000139617), score: -0.49 NAGKN-acetylglucosamine kinase (ENSG00000124357), score: 0.56 NAT10N-acetyltransferase 10 (GCN5-related) (ENSG00000135372), score: -0.55 NBEAL1neurobeachin-like 1 (ENSG00000144426), score: 0.48 NDUFB9NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa (ENSG00000147684), score: -0.61 NEIL2nei endonuclease VIII-like 2 (E. coli) (ENSG00000154328), score: 0.5 NFIL3nuclear factor, interleukin 3 regulated (ENSG00000165030), score: 0.49 NLRP12NLR family, pyrin domain containing 12 (ENSG00000142405), score: 0.47 NOP2NOP2 nucleolar protein homolog (yeast) (ENSG00000111641), score: 0.47 NQO1NAD(P)H dehydrogenase, quinone 1 (ENSG00000181019), score: 0.45 NRLneural retina leucine zipper (ENSG00000129535), score: -0.52 NT5C3L5'-nucleotidase, cytosolic III-like (ENSG00000141698), score: -0.52 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (ENSG00000170917), score: 0.44 NXF1nuclear RNA export factor 1 (ENSG00000162231), score: 0.45 OR1D2olfactory receptor, family 1, subfamily D, member 2 (ENSG00000184166), score: 0.53 OSGEPL1O-sialoglycoprotein endopeptidase-like 1 (ENSG00000128694), score: 0.51 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.45 PAPD7PAP associated domain containing 7 (ENSG00000112941), score: -0.55 PARP3poly (ADP-ribose) polymerase family, member 3 (ENSG00000041880), score: 0.44 PCYT1Aphosphate cytidylyltransferase 1, choline, alpha (ENSG00000161217), score: 0.45 PDCD4programmed cell death 4 (neoplastic transformation inhibitor) (ENSG00000150593), score: 0.5 PFDN6prefoldin subunit 6 (ENSG00000204220), score: -0.98 PGLYRP1peptidoglycan recognition protein 1 (ENSG00000008438), score: 0.48 PHF23PHD finger protein 23 (ENSG00000040633), score: 0.97 PIM2pim-2 oncogene (ENSG00000102096), score: 0.52 PLA2G16phospholipase A2, group XVI (ENSG00000176485), score: 0.56 PLA2G2Fphospholipase A2, group IIF (ENSG00000158786), score: 0.52 PLAC8placenta-specific 8 (ENSG00000145287), score: 0.46 PLEKHG2pleckstrin homology domain containing, family G (with RhoGef domain) member 2 (ENSG00000090924), score: 0.55 PLTPphospholipid transfer protein (ENSG00000100979), score: -0.55 PM20D2peptidase M20 domain containing 2 (ENSG00000146281), score: 0.54 PODNpodocan (ENSG00000174348), score: 0.51 POLE4polymerase (DNA-directed), epsilon 4 (p12 subunit) (ENSG00000115350), score: 0.47 POLR1Epolymerase (RNA) I polypeptide E, 53kDa (ENSG00000137054), score: 0.51 POLR2Fpolymerase (RNA) II (DNA directed) polypeptide F (ENSG00000100142), score: -0.69 POLR2Ipolymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa (ENSG00000105258), score: -0.66 POMCproopiomelanocortin (ENSG00000115138), score: -0.48 POU2F3POU class 2 homeobox 3 (ENSG00000137709), score: 0.55 PPA1pyrophosphatase (inorganic) 1 (ENSG00000180817), score: -0.52 PPBPpro-platelet basic protein (chemokine (C-X-C motif) ligand 7) (ENSG00000163736), score: 0.58 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta (ENSG00000175470), score: -0.49 PRICKLE4prickle homolog 4 (Drosophila) (ENSG00000124593), score: -0.53 PRLprolactin (ENSG00000172179), score: 0.7 PRMT3protein arginine methyltransferase 3 (ENSG00000185238), score: 0.52 PROSCproline synthetase co-transcribed homolog (bacterial) (ENSG00000147471), score: 0.47 PRSS53protease, serine, 53 (ENSG00000151006), score: 0.55 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (ENSG00000204264), score: -0.64 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (ENSG00000240065), score: -0.7 PTCD3Pentatricopeptide repeat domain 3 (ENSG00000132300), score: 0.71 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: 0.51 PYCR1pyrroline-5-carboxylate reductase 1 (ENSG00000183010), score: 0.5 R3HDMLR3H domain containing-like (ENSG00000101074), score: 0.5 RAB40ARAB40A, member RAS oncogene family (ENSG00000172476), score: 0.56 RABGGTBRab geranylgeranyltransferase, beta subunit (ENSG00000137955), score: 0.47 RAD9ARAD9 homolog A (S. pombe) (ENSG00000172613), score: -0.5 RARSarginyl-tRNA synthetase (ENSG00000113643), score: 0.49 RBM12BRNA binding motif protein 12B (ENSG00000183808), score: 0.46 RBX1ring-box 1 (ENSG00000100387), score: 0.45 RFC2replication factor C (activator 1) 2, 40kDa (ENSG00000049541), score: 0.48 RFX8regulatory factor X, 8 (ENSG00000196460), score: 0.69 RGL2ral guanine nucleotide dissociation stimulator-like 2 (ENSG00000237441), score: -0.96 RGS2regulator of G-protein signaling 2, 24kDa (ENSG00000116741), score: 0.44 RING1ring finger protein 1 (ENSG00000204227), score: -1 RNASE7ribonuclease, RNase A family, 7 (ENSG00000165799), score: 0.46 RNLSrenalase, FAD-dependent amine oxidase (ENSG00000184719), score: 0.52 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: -0.7 RPL37ribosomal protein L37 (ENSG00000145592), score: -0.85 RPUSD3RNA pseudouridylate synthase domain containing 3 (ENSG00000156990), score: -0.55 RRP1ribosomal RNA processing 1 homolog (S. cerevisiae) (ENSG00000160214), score: 0.45 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (ENSG00000067533), score: 0.6 RXRBretinoid X receptor, beta (ENSG00000204231), score: -0.99 S100A12S100 calcium binding protein A12 (ENSG00000163221), score: 0.51 SAP18Sin3A-associated protein, 18kDa (ENSG00000150459), score: 0.61 SAP30BPSAP30 binding protein (ENSG00000161526), score: 0.57 SCARF1scavenger receptor class F, member 1 (ENSG00000074660), score: 0.47 SCGB3A2secretoglobin, family 3A, member 2 (ENSG00000164265), score: 0.54 SCO1SCO cytochrome oxidase deficient homolog 1 (yeast) (ENSG00000133028), score: 0.44 SCYL1SCY1-like 1 (S. cerevisiae) (ENSG00000142186), score: 0.95 SDCBPsyndecan binding protein (syntenin) (ENSG00000137575), score: 0.51 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.55 SERBP1SERPINE1 mRNA binding protein 1 (ENSG00000142864), score: 0.53 SETMARSET domain and mariner transposase fusion gene (ENSG00000170364), score: 0.5 SFT2D1SFT2 domain containing 1 (ENSG00000198818), score: -0.54 SFXN2sideroflexin 2 (ENSG00000156398), score: 0.49 SLC30A5solute carrier family 30 (zinc transporter), member 5 (ENSG00000145740), score: 0.44 SLC39A7solute carrier family 39 (zinc transporter), member 7 (ENSG00000112473), score: -0.97 SLC44A2solute carrier family 44, member 2 (ENSG00000129353), score: -0.48 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: -0.49 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: 0.59 SNRNP48small nuclear ribonucleoprotein 48kDa (U11/U12) (ENSG00000168566), score: 0.58 SRFBP1serum response factor binding protein 1 (ENSG00000151304), score: 0.63 SRP19signal recognition particle 19kDa (ENSG00000153037), score: 0.45 SRSF3serine/arginine-rich splicing factor 3 (ENSG00000112081), score: 0.47 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: 0.5 SSBP1single-stranded DNA binding protein 1 (ENSG00000106028), score: 0.48 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: 0.62 SYF2SYF2 homolog, RNA splicing factor (S. cerevisiae) (ENSG00000117614), score: 0.46 TAP1transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) (ENSG00000168394), score: -0.76 TAPBPTAP binding protein (tapasin) (ENSG00000231925), score: -0.94 TBC1D14TBC1 domain family, member 14 (ENSG00000132405), score: -0.53 TCF19transcription factor 19 (ENSG00000137310), score: -0.59 TCTE3t-complex-associated-testis-expressed 3 (ENSG00000184786), score: 0.51 TCTEX1D1Tctex1 domain containing 1 (ENSG00000152760), score: 0.61 TFF1trefoil factor 1 (ENSG00000160182), score: 0.59 THOC5THO complex 5 (ENSG00000100296), score: 0.44 THOC7THO complex 7 homolog (Drosophila) (ENSG00000163634), score: 0.48 THRAP3thyroid hormone receptor associated protein 3 (ENSG00000054118), score: 0.48 THTPAthiamine triphosphatase (ENSG00000157306), score: -0.51 THUMPD3THUMP domain containing 3 (ENSG00000134077), score: -0.81 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (ENSG00000151923), score: 0.53 TIAM2T-cell lymphoma invasion and metastasis 2 (ENSG00000146426), score: -0.47 TIGITT cell immunoreceptor with Ig and ITIM domains (ENSG00000181847), score: 0.49 TMC3transmembrane channel-like 3 (ENSG00000188869), score: 0.55 TMCO1transmembrane and coiled-coil domains 1 (ENSG00000143183), score: 0.54 TMEM101transmembrane protein 101 (ENSG00000091947), score: -0.54 TMEM131transmembrane protein 131 (ENSG00000075568), score: -0.62 TMEM149transmembrane protein 149 (ENSG00000126246), score: 0.5 TMEM187transmembrane protein 187 (ENSG00000177854), score: -0.54 TMEM80transmembrane protein 80 (ENSG00000177042), score: -0.66 TRAT1T cell receptor associated transmembrane adaptor 1 (ENSG00000163519), score: 0.5 TRDMT1tRNA aspartic acid methyltransferase 1 (ENSG00000107614), score: 0.5 TRIM26tripartite motif-containing 26 (ENSG00000234127), score: -0.95 TRIM5tripartite motif-containing 5 (ENSG00000132256), score: 0.45 TRIT1tRNA isopentenyltransferase 1 (ENSG00000043514), score: 0.66 TRNT1tRNA nucleotidyl transferase, CCA-adding, 1 (ENSG00000072756), score: 0.49 TRUB2TruB pseudouridine (psi) synthase homolog 2 (E. coli) (ENSG00000167112), score: 0.48 TTC14tetratricopeptide repeat domain 14 (ENSG00000163728), score: 0.45 TXNDC12thioredoxin domain containing 12 (endoplasmic reticulum) (ENSG00000117862), score: 0.48 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (ENSG00000197355), score: 0.45 UBA5ubiquitin-like modifier activating enzyme 5 (ENSG00000081307), score: 0.45 UBE2Bubiquitin-conjugating enzyme E2B (RAD6 homolog) (ENSG00000119048), score: 0.5 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: 0.53 UBE2Iubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) (ENSG00000103275), score: -0.49 UBE3Cubiquitin protein ligase E3C (ENSG00000009335), score: -0.46 UBR4ubiquitin protein ligase E3 component n-recognin 4 (ENSG00000127481), score: -0.47 UPK1Auroplakin 1A (ENSG00000105668), score: 0.46 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (ENSG00000142207), score: -0.54 UTP14AUTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) (ENSG00000156697), score: 0.58 UTP23UTP23, small subunit (SSU) processome component, homolog (yeast) (ENSG00000147679), score: 0.5 VAMP7vesicle-associated membrane protein 7 (ENSG00000124333), score: 0.46 VILLvillin-like (ENSG00000136059), score: 0.47 VMO1vitelline membrane outer layer 1 homolog (chicken) (ENSG00000182853), score: 0.53 VNN2vanin 2 (ENSG00000112303), score: 0.61 VPS52vacuolar protein sorting 52 homolog (S. cerevisiae) (ENSG00000223501), score: -1 WBP1WW domain binding protein 1 (ENSG00000115274), score: -0.51 WBP5WW domain binding protein 5 (ENSG00000185222), score: 0.56 WDR46WD repeat domain 46 (ENSG00000227057), score: -0.99 WDR83WD repeat domain 83 (ENSG00000123154), score: -0.51 WFDC3WAP four-disulfide core domain 3 (ENSG00000124116), score: 0.46 WRBtryptophan rich basic protein (ENSG00000182093), score: -0.53 ZBTB22zinc finger and BTB domain containing 22 (ENSG00000236104), score: -0.95 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: 0.46 ZC3H3zinc finger CCCH-type containing 3 (ENSG00000014164), score: -0.47 ZCCHC14zinc finger, CCHC domain containing 14 (ENSG00000140948), score: -0.49 ZCCHC4zinc finger, CCHC domain containing 4 (ENSG00000168228), score: 0.5 ZDHHC8zinc finger, DHHC-type containing 8 (ENSG00000099904), score: -0.52 ZMAT1zinc finger, matrin type 1 (ENSG00000166432), score: 0.47 ZNF232zinc finger protein 232 (ENSG00000167840), score: 0.65 ZNF275zinc finger protein 275 (ENSG00000063587), score: -0.51 ZNF343zinc finger protein 343 (ENSG00000088876), score: -0.46 ZNF394zinc finger protein 394 (ENSG00000160908), score: 0.62 ZNF434zinc finger protein 434 (ENSG00000140987), score: -0.51 ZNF470zinc finger protein 470 (ENSG00000197016), score: -0.47 ZNF485zinc finger protein 485 (ENSG00000198298), score: 0.47 ZNF582zinc finger protein 582 (ENSG00000018869), score: 0.5 ZNF584zinc finger protein 584 (ENSG00000171574), score: -0.55 ZNF589zinc finger protein 589 (ENSG00000164048), score: -0.78 ZNF629zinc finger protein 629 (ENSG00000102870), score: -0.62 ZNF652zinc finger protein 652 (ENSG00000198740), score: 0.54 ZNF692zinc finger protein 692 (ENSG00000171163), score: 0.45 ZNF707zinc finger protein 707 (ENSG00000181135), score: -0.51 ZNF792zinc finger protein 792 (ENSG00000180884), score: 0.5 ZNF83zinc finger protein 83 (ENSG00000167766), score: 0.47 ZNRD1zinc ribbon domain containing 1 (ENSG00000066379), score: -0.9 ZSCAN1zinc finger and SCAN domain containing 1 (ENSG00000152467), score: -0.46 ZZEF1zinc finger, ZZ-type with EF-hand domain 1 (ENSG00000074755), score: -0.5
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| ppy_br_m_ca1 | ppy | br | m | _ |
| ppy_cb_f_ca1 | ppy | cb | f | _ |
| ppy_br_f_ca1 | ppy | br | f | _ |
| ppy_kd_m_ca1 | ppy | kd | m | _ |
| ppy_kd_f_ca1 | ppy | kd | f | _ |
| ppy_ht_m_ca1 | ppy | ht | m | _ |
| ppy_ht_f_ca1 | ppy | ht | f | _ |
| ppy_lv_m_ca1 | ppy | lv | m | _ |
| ppy_lv_f_ca1 | ppy | lv | f | _ |